Structural insight into DNA-assembled oligochromophores: crystallographic analysis of pyrene- and phenanthrene-modified DNA in complex with BpuJI endonuclease.
Probst, M., Aeschimann, W., Chau, T.T., Langenegger, S.M., Stocker, A., Haner, R.(2016) Nucleic Acids Res 44: 7079-7089
- PubMed: 27422870
- DOI: https://doi.org/10.1093/nar/gkw644
- Primary Citation of Related Structures:
5HLT, 5HNF, 5HNH - PubMed Abstract:
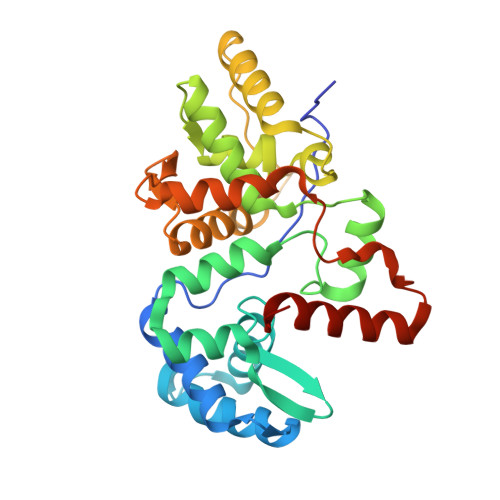
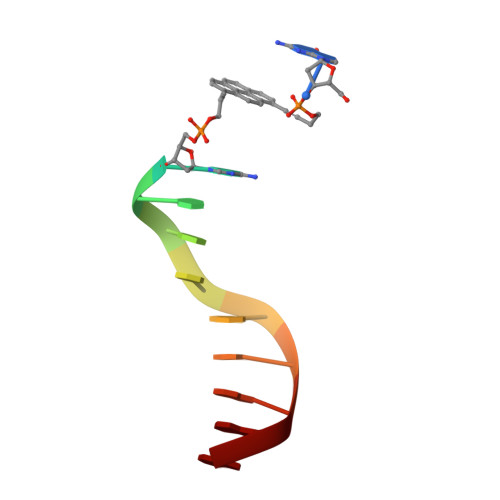
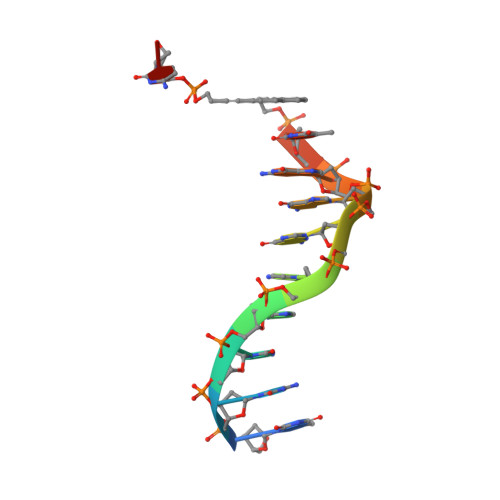
The use of the DNA duplex as a supramolecular scaffold is an established approach for the assembly of chromophore aggregates. In the absence of detailed structural insight, the characterization of thus assembled oligochromophores is, today, largely based on solution-phase spectroscopy. Here, we describe the crystal structures of three DNA-organized chromophore aggregates. DNA hybrids containing non-nucleosidic pyrene and phenanthrene building blocks were co-crystallized with the recently described binding domain of the restriction enzyme BpuJI. Crystal structures of these complexes were determined at 2.7, 1.9 and 1.6 Å resolutions. The structures reveal aromatic stacking interactions between pyrene and/or phenanthrene units within the framework of the B-DNA duplex. In hybrids containing a single modification in each DNA strand near the end of the duplex, the two polyaromatic hydrocarbons are engaged in a face-to-face stacking orientation. Due to crystal packing and steric effects, the terminal GC base pair is disrupted in all three crystal structures, which results in a non-perfect stacking arrangement of the aromatic chromophores in two of the structures. In a hybrid containing a total of three pyrenes, crystal lattice induced end-to-end stacking of individual DNA duplexes leads to the formation of an extended aromatic π-stack containing four co-axially arranged pyrenes. The aromatic planes of the stacked pyrenes are oriented in a parallel way. The study demonstrates the value of co-crystallization of chemically modified DNA with the recombinant binding domain of the restriction enzyme BpuJI for obtaining detailed structural insight into DNA-assembled oligochromophores.
- Department of Chemistry and Biochemistry, University of Bern, 3012 Bern, Switzerland.
Organizational Affiliation: