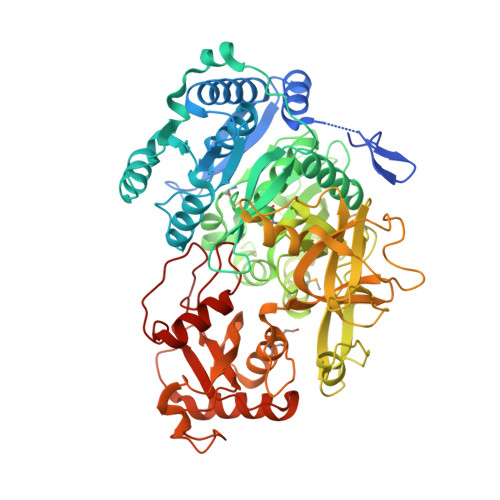
Structure of the Essential Mtb FadD32 Enzyme: A Promising Drug Target for Treating Tuberculosis.
Kuhn, M.L., Alexander, E., Minasov, G., Page, H.J., Warwrzak, Z., Shuvalova, L., Flores, K.J., Wilson, D.J., Shi, C., Aldrich, C.C., Anderson, W.F.(2016) ACS Infect Dis 2: 579-591
- PubMed: 27547819
- DOI: https://doi.org/10.1021/acsinfecdis.6b00082
- Primary Citation of Related Structures:
5HM3 - PubMed Abstract:
Mycolic acids are indispensible lipids of Mycobacterium tuberculosis ( Mtb ), the causative agent of tuberculosis (TB), and contribute to the distinctive architecture and impermeability of the mycobacterial cell envelope. FadD32 plays a pivotal role in mycolic acid biosynthesis by functionally linking fatty acid synthase (FAS) and polyketide synthase (PKS) biosynthetic pathways. FadD32, a fatty acyl-AMP ligase (FAAL), represents one of the best genetically and chemically validated new TB drug targets. We have determined the three-dimensional crystal structure of Mtb FadD32 in complex with a ligand specifically designed to stabilize the catalytically active adenylate-conformation, which provides a foundation for structure-based drug design efforts against this essential protein. The structure also captures the unique interactions of a FAAL-specific insertion sequence and provides insight into the specificity and mechanism of fatty acid transfer.
- Center for Structural Genomics of Infectious Diseases, Department of Biochemistry and Molecular Genetics, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, United States; Center for Drug Design, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Organizational Affiliation: