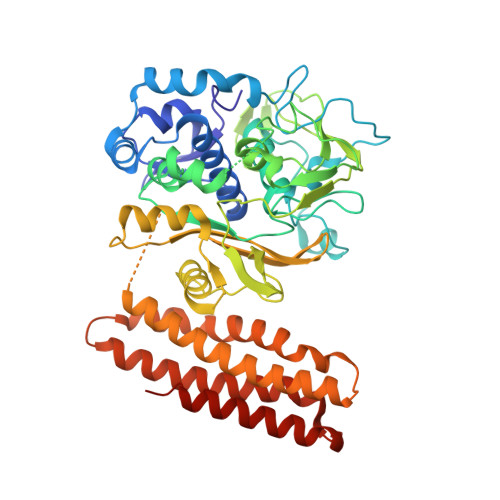
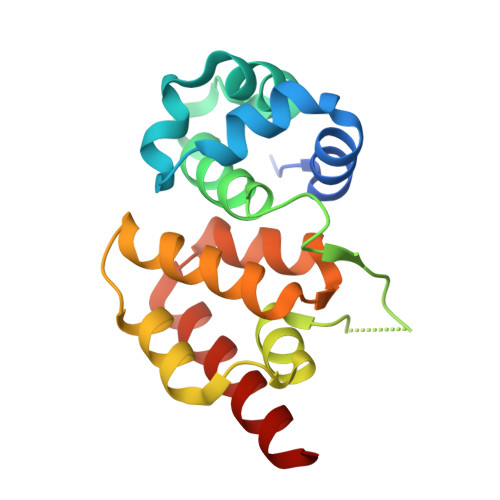
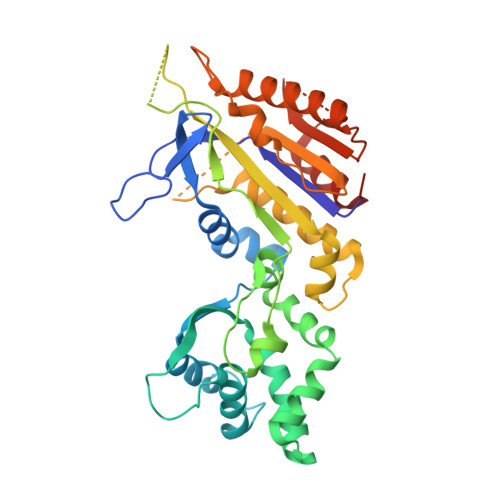
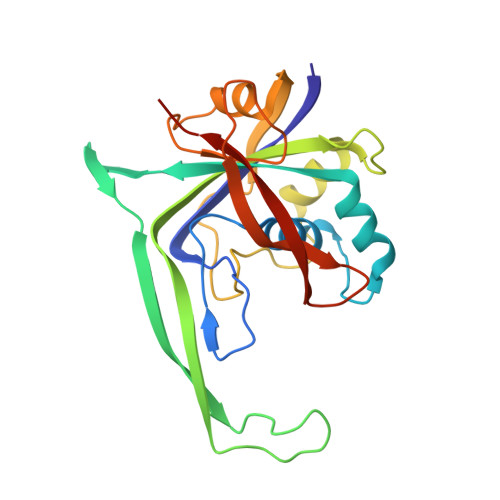
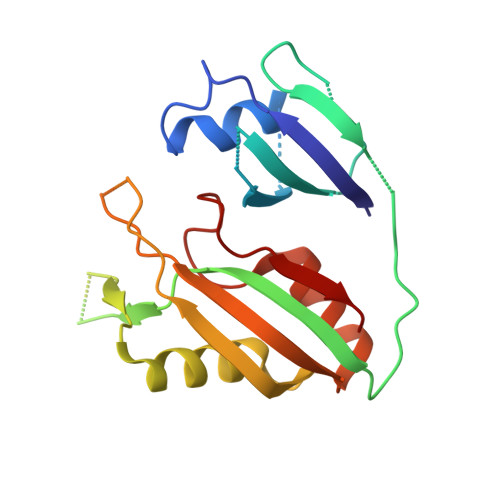
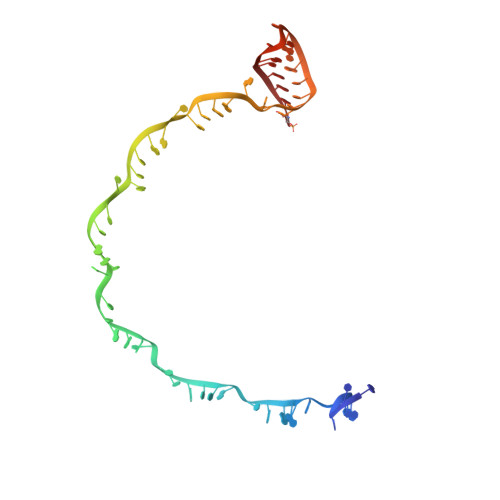
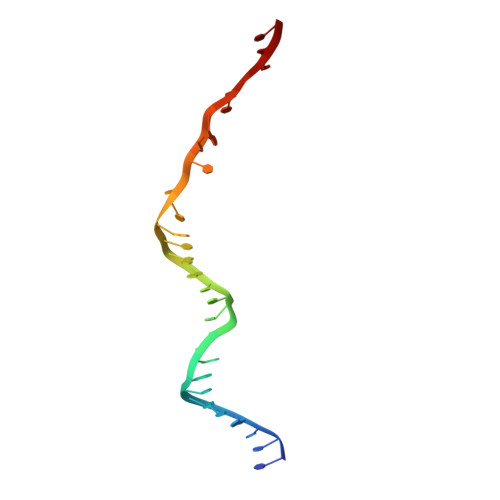
Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Hayes, R.P., Xiao, Y., Ding, F., van Erp, P.B., Rajashankar, K., Bailey, S., Wiedenheft, B., Ke, A.(2016) Nature 530: 499-503
- PubMed: 26863189
- DOI: https://doi.org/10.1038/nature16995
- Primary Citation of Related Structures:
5H9E, 5H9F - PubMed Abstract:
Clustered regularly interspaced short palindromic repeats (CRISPRs) and the cas (CRISPR-associated) operon form an RNA-based adaptive immune system against foreign genetic elements in prokaryotes. Type I accounts for 95% of CRISPR systems, and has been used to control gene expression and cell fate. During CRISPR RNA (crRNA)-guided interference, Cascade (CRISPR-associated complex for antiviral defence) facilitates the crRNA-guided invasion of double-stranded DNA for complementary base-pairing with the target DNA strand while displacing the non-target strand, forming an R-loop. Cas3, which has nuclease and helicase activities, is subsequently recruited to degrade two DNA strands. A protospacer adjacent motif (PAM) sequence flanking target DNA is crucial for self versus foreign discrimination. Here we present the 2.45 Å crystal structure of Escherichia coli Cascade bound to a foreign double-stranded DNA target. The 5'-ATG PAM is recognized in duplex form, from the minor groove side, by three structural features in the Cascade Cse1 subunit. The promiscuity inherent to minor groove DNA recognition rationalizes the observation that a single Cascade complex can respond to several distinct PAM sequences. Optimal PAM recognition coincides with wedge insertion, initiating directional target DNA strand unwinding to allow segmented base-pairing with crRNA. The non-target strand is guided along a parallel path 25 Å apart, and the R-loop structure is further stabilized by locking this strand behind the Cse2 dimer. These observations provide the structural basis for understanding the PAM-dependent directional R-loop formation process.
- Department of Molecular Biology and Genetics, Cornell University, 253 Biotechnology Building, Ithaca, New York 14853, USA.
Organizational Affiliation: