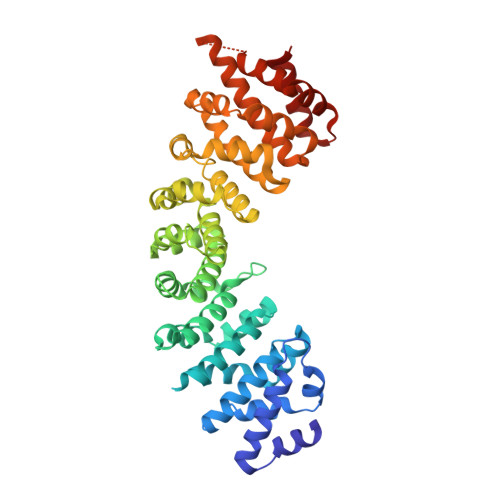
Structures of the Karyopherins Kap121p and Kap60p Bound to the Nuclear Pore-Targeting Domain of the SUMO Protease Ulp1p
Hirano, H., Kobayashi, J., Matsuura, Y.(2017) J Mol Biology 429: 249-260
- PubMed: 27939291
- DOI: https://doi.org/10.1016/j.jmb.2016.11.029
- Primary Citation of Related Structures:
5H2V, 5H2W, 5H2X - PubMed Abstract:
The budding yeast small ubiquitin-like modifier (SUMO) protease Ulp1p catalyzes both the processing of newly synthesized SUMO to its mature form and the deconjugation of SUMO from target proteins, thereby regulating a wide range of cellular processes including cell division, DNA repair, DNA replication, transcription, and mRNA quality control. Ulp1p is localized primarily at the nuclear pore complex (NPC) through interactions involving the karyopherins Kap121p and Kap95p-Kap60p heterodimer and a subset of nuclear pore-associated proteins. The sequestration of Ulp1p at the nuclear periphery is crucial for the proper control of protein desumoylation. To gain insights into the role of the karyopherins in regulating the localization of Ulp1p, we have determined the crystal structures of Kap121p and Kap60p bound to the N-terminal non-catalytic domain of Ulp1p that is necessary and sufficient for NPC targeting. Contrary to a previous proposal that Ulp1p is tethered to the transport channel of the NPC through unconventional interactions with the karyopherins, our structures reveal that Ulp1p has canonical nuclear localization signals (NLSs): (1) an isoleucine-lysine-NLS (residues 51-55) that binds to the NLS-binding site of Kap121p, and (2) a classical bipartite NLS (residues 154-172) that binds to the major and minor NLS-binding sites of Kap60p. Ulp1p also binds Kap95p directly, and the Ulp1p-Kap95p binding is enhanced by the importin-β-binding domain of Kap60p. GTP-bound Gsp1p (the yeast Ran ortholog) and the exportin Cse1p cooperate to release Ulp1p from the karyopherins, indicating that the stable sequestration of Ulp1p to the NPC would require a karyopherin-independent mechanism to anchor Ulp1p at the NPC.
- Division of Biological Science, Graduate School of Science, Nagoya University, 464-8602, Japan; Structural Biology Research Center, Graduate School of Science, Nagoya University, 464-8602, Japan.
Organizational Affiliation: