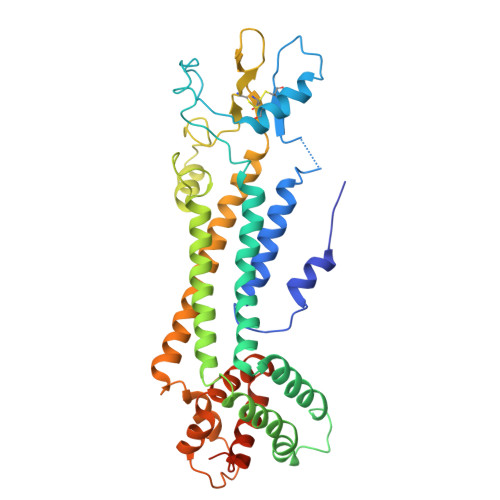
Atomic structure of the innexin-6 gap junction channel determined by cryo-EM
Oshima, A., Tani, K., Fujiyoshi, Y.(2016) Nat Commun 7: 13681-13681
- PubMed: 27905396
- DOI: https://doi.org/10.1038/ncomms13681
- Primary Citation of Related Structures:
5H1Q, 5H1R - PubMed Abstract:
Innexins, a large protein family comprising invertebrate gap junction channels, play an essential role in nervous system development and electrical synapse formation. Here we report the cryo-electron microscopy structures of Caenorhabditis elegans innexin-6 (INX-6) gap junction channels at atomic resolution. We find that the arrangements of the transmembrane helices and extracellular loops of the INX-6 monomeric structure are highly similar to those of connexin-26 (Cx26), despite the lack of significant sequence similarity. The INX-6 gap junction channel comprises hexadecameric subunits but reveals the N-terminal pore funnel, consistent with Cx26. The helix-rich cytoplasmic loop and C-terminus are intercalated one-by-one through an octameric hemichannel, forming a dome-like entrance that interacts with N-terminal loops in the pore. These observations suggest that the INX-6 cytoplasmic domains are cooperatively associated with the N-terminal funnel conformation, and an essential linkage of the N-terminal with channel activity is presumably preserved across gap junction families.
- Cellular and Structural Physiology Institute (CeSPI), Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8601, Japan.
Organizational Affiliation: