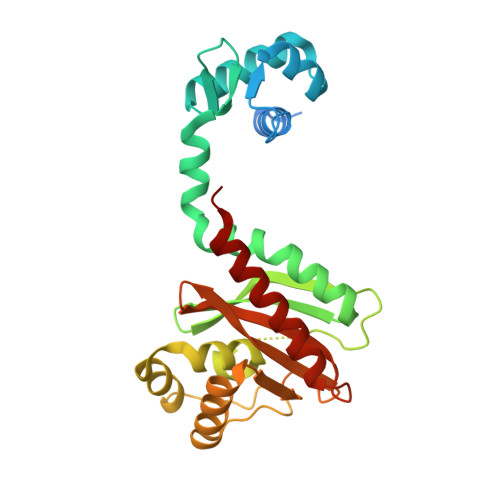
Crystal structure of an IclR homologue from Microbacterium sp. strain HM58-2.
Akiyama, T., Yamada, Y., Takaya, N., Ito, S., Sasaki, Y., Yajima, S.(2017) Acta Crystallogr F Struct Biol Commun 73: 16-23
- PubMed: 28045389
- DOI: https://doi.org/10.1107/S2053230X16019208
- Primary Citation of Related Structures:
5H1A - PubMed Abstract:
The bacterial transcription factor IclR (isocitrate lyase regulator) is a member of a one-component signal transduction system, which shares the common motif of a helix-turn-helix (HTH)-type DNA-binding domain (DBD) connected to a substrate-binding domain (SBD). Here, the crystal structure of an IclR homologue (Mi-IclR) from Microbacterium sp. strain HM58-2, which catabolizes acylhydrazide as the sole carbon source, is reported. Mi-IclR is expected to regulate an operon responsible for acylhydrazide degradation as an initial step. Native single-wavelength anomalous diffraction (SAD) experiments were performed in combination with molecular replacement. CRANK2 from the CCP4 suite successfully phased and modelled the complete structure of a homotetramer composed of 1000 residues in an asymmetric unit, and the model was refined to 2.1 Å resolution. The overall structure of Mi-IclR shared the same domain combination as other known IclR structures, but the relative geometry between the DBD and SBD differs. Accordingly, the geometry of the Mi-IclR tetramer was unique: the putative substrate-binding site in each subunit is accessible from the outside of the tetramer, as opposed to buried inside as in the previously known IclR structures. These differences in the domain geometry may contribute to the transcriptional regulation of IclRs.
- Department of Bioscience, Tokyo University of Agriculture, Setagaya-ku, Tokyo 156-8502, Japan.
Organizational Affiliation: