Michael Acceptor-Based Peptidomimetic Inhibitor of Main Protease from Porcine Epidemic Diarrhea Virus
Wang, F., Chen, C., Yang, K., Xu, Y., Liu, X., Gao, F., Liu, H., Chen, X., Zhao, Q., Liu, X., Cai, Y., Yang, H.(2017) J Med Chem 60: 3212-3216
- PubMed: 28287727
- DOI: https://doi.org/10.1021/acs.jmedchem.7b00103
- Primary Citation of Related Structures:
5GWZ - PubMed Abstract:
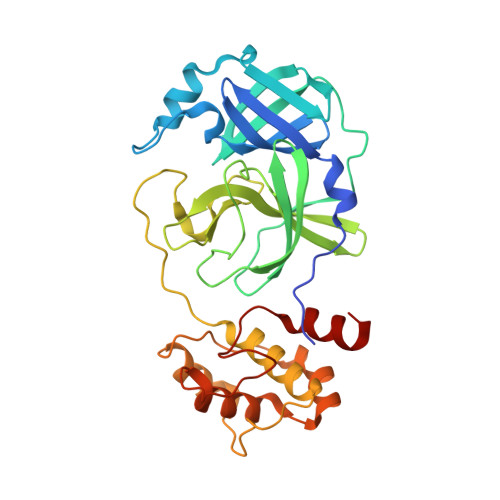
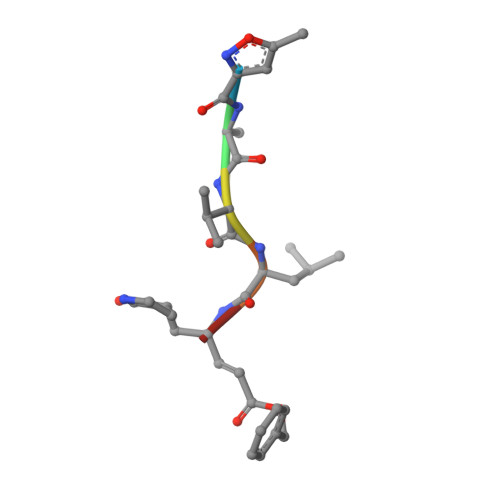
Porcine epidemic diarrhea virus (PEDV) causes high mortality in pigs. PEDV main protease (M pro ) plays an essential role in viral replication. We solved the structure of PEDV M pro complexed with peptidomimetic inhibitor N3 carrying a Michael acceptor warhead, revealing atomic level interactions. We further designed a series of 17 inhibitors with altered side groups. Inhibitors M2 and M17 demonstrated enhanced specificity against PEDV M pro . These compounds have potential as future therapeutics to combat PEDV infection.
- School of Life Sciences, Tianjin University , Tianjin 300072, China.
Organizational Affiliation: