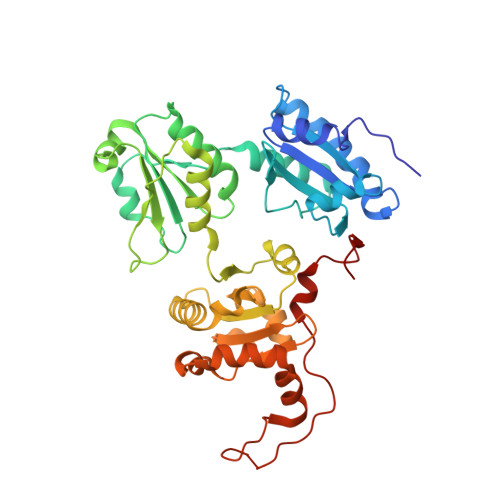
Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface
Watanabe, S., Harayama, M., Kanemura, S., Sitia, R., Inaba, K.(2017) Proc Natl Acad Sci U S A 114: E3224-E3232
- PubMed: 28373561
- DOI: https://doi.org/10.1073/pnas.1621426114
- Primary Citation of Related Structures:
5GU6, 5GU7 - PubMed Abstract:
ERp44 retrieves some endoplasmic reticulum (ER)-resident enzymes and immature oligomers of secretory proteins from the Golgi. Association of ERp44 with its clients is regulated by pH-dependent mechanisms, but the molecular details are not fully understood. Here we report high-resolution crystal structures of human ERp44 at neutral and weakly acidic pH. These structures reveal key regions in the C-terminal tail (C tail) missing in the original crystal structure, including a regulatory histidine-rich region and a subsequent extended loop. The former region forms a short α-helix (α16), generating a histidine-clustered site (His cluster). At low pH, the three Trx-like domains of ERp44 ("a," "b," and "b'") undergo significant rearrangements, likely induced by protonation of His157 located at the interface between the a and b domains. The α16-helix is partially unwound and the extended loop is disordered in weakly acidic conditions, probably due to electrostatic repulsion between the protonated histidines in the His cluster. Molecular dynamics simulations indicated that helix unwinding enhances the flexibility of the C tail, disrupting its normal hydrogen-bonding pattern. The observed pH-dependent conformational changes significantly enlarge the positively charged regions around the client-binding site of ERp44 at low pH. Mutational analyses showed that ERp44 forms mixed disulfides with specific cysteines residing on negatively charged loop regions of Ero1α. We propose that the protonation states of the essential histidines regulate the ERp44-client interaction by altering the C-tail dynamics and surface electrostatic potential of ERp44.
- Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Sendai 980-8577, Japan.
Organizational Affiliation: