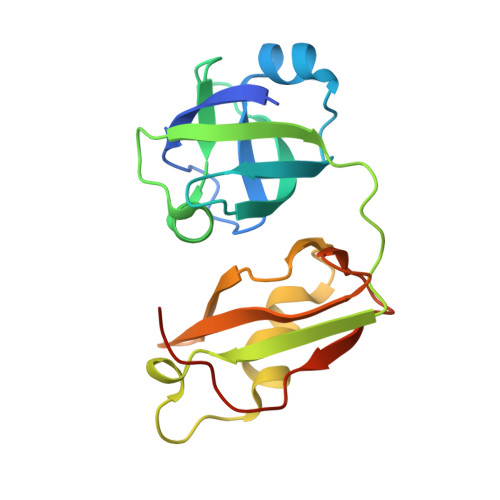
Structural insights into the interaction of human p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease.
Lim, J.J., Lee, Y., Yoon, S.Y., Ly, T.T., Kang, J.Y., Youn, H.S., An, J.Y., Lee, J.G., Park, K.R., Kim, T.G., Yang, J.K., Jun, Y., Eom, S.H.(2016) FEBS Lett 590: 4402-4413
- PubMed: 27714797
- DOI: https://doi.org/10.1002/1873-3468.12447
- Primary Citation of Related Structures:
5GLF - PubMed Abstract:
The interaction of the rhomboid pseudoprotease Derlin-1 and p97 is crucial for the retrotranslocation of polyubiquitinated substrates in the endoplasmic reticulum-associated degradation pathway. We report a 2.25 Å resolution structure of the p97 N-terminal domain (p97N) in complex with the Derlin-1 SHP motif. Remarkably, the SHP motif adopts a short, antiparallel β-strand that interacts with the β-sheet of p97N-a site distinct from that to which most p97 adaptor proteins bind. Mutational and biochemical analyses contributed to defining the specific interaction, demonstrating the importance of a highly conserved binding pocket on p97N and a signature motif on SHP. Our findings may also provide insights into the interactions between other SHP-containing proteins and p97N.
- School of Life Science, Gwangju Institute of Science and Technology (GIST), Korea.
Organizational Affiliation: