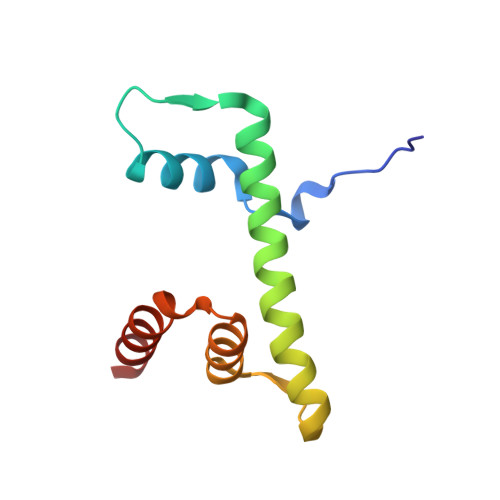
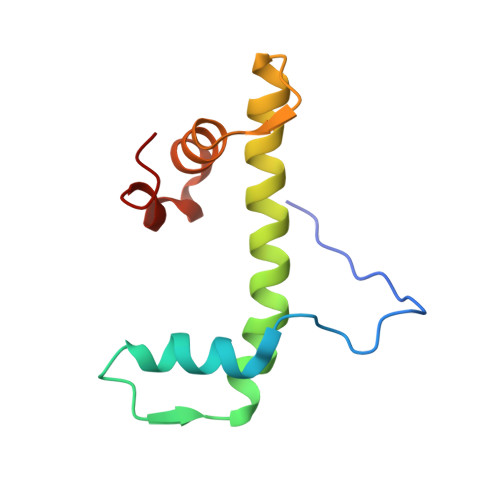
Crystal Structure of the Arabidopsis thaliana L1L/NF-YC3 Histone-fold Dimer Reveals Specificities of the LEC1 Family of NF-Y Subunits in Plants.
Gnesutta, N., Saad, D., Chaves-Sanjuan, A., Mantovani, R., Nardini, M.(2017) Mol Plant 10: 645-648
- PubMed: 27871811
- DOI: https://doi.org/10.1016/j.molp.2016.11.006
- Primary Citation of Related Structures:
5G49 - Dipartimento di Bioscienze, Università degli Studi di Milano, Via Celoria 26, 20133 Milan, Italy.
Organizational Affiliation: