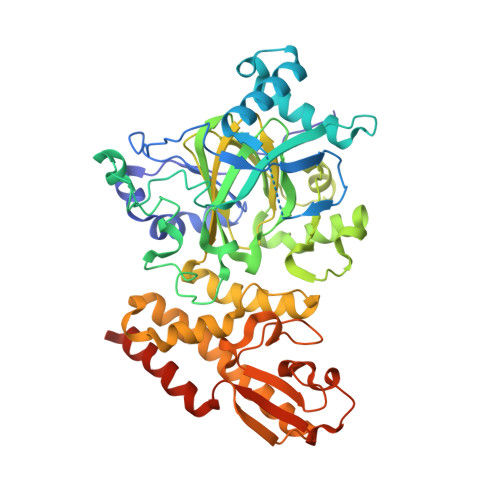
Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Malate
Nowak, R., Krojer, T., Johansson, C., Gileadi, C., Kupinska, K., Strain-Damerell, C., Szykowska, A., von Delft, F., Burgess-Brown, N.A., Arrowsmith, C.H., Bountra, C., Edwards, A.M., Oppermann, U.To be published.