Chemoenzymatic Synthesis of 6-Phospho-Cyclophellitol as a Novel Probe of 6-Phospho-Beta-Glucosidases.
Kwan, D.H., Jin, Y., Jiang, J., Chen, H., Kotzler, M.P., Overkleeft, H.S., Davies, G.J., Withers, S.G.(2016) FEBS Lett 590: 461
- PubMed: 26790390
- DOI: https://doi.org/10.1002/1873-3468.12059
- Primary Citation of Related Structures:
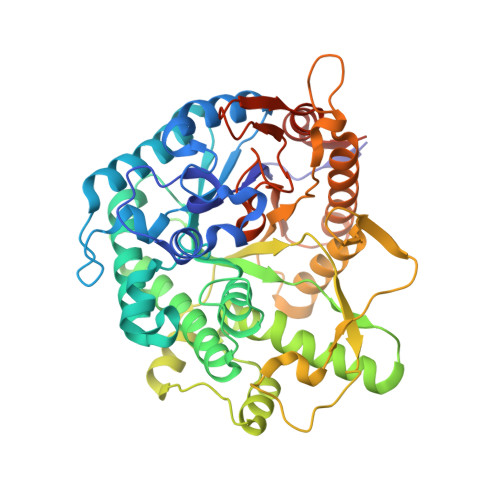
5FOO - PubMed Abstract:
Covalent, mechanism-based inhibitors of glycosidases are valuable probe molecules for visualizing enzyme activities in complex systems. We, here, describe the chemoenzymatic synthesis of 6-phospho-cyclophellitol and evaluate its behaviour as a mechanism-based inactivator of the Streptococcus pyogenes 6-phospho-β-glucosidase from CAZy family GH1. We further present the three-dimensional structure of the inactivated enzyme, which reveals the constellation of active site residues responsible for the enzyme's specificity and confirms the covalent nature of the inactivation.
- Department of Chemistry, University of British Columbia, BC, Canada.
Organizational Affiliation: