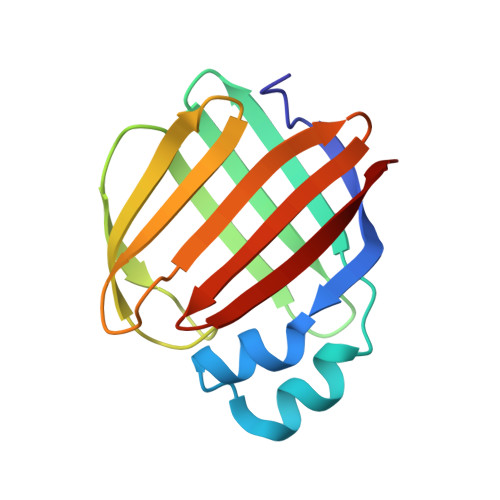
Crystal structure of the Q108K:K40L:T51V:R58W:Y19W mutant of human Cellular Retinol Binding ProteinII in complex with All-trans-Retinal at 1.68 Angstrom Resolution
Nossoni, Z., Nosrati, M., Wang, W., Berbasova, T., Vasileiou, C., Borhan, B., Geiger, J.H.To be published.