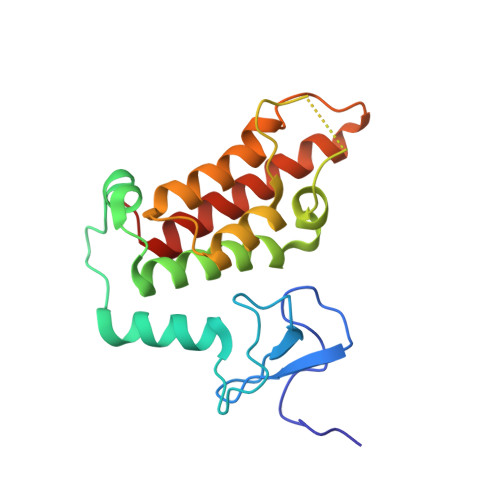
Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C
Zhang, X., Zhao, D., Xiong, X., He, Z., Li, H.(2016) J Biological Chem 291: 12786-12798
- PubMed: 27129259
- DOI: https://doi.org/10.1074/jbc.M116.721159
- Primary Citation of Related Structures:
5FB0, 5FB1 - PubMed Abstract:
The decoding of histone post-translational modifications by chromatin-binding modules ("readers") constitutes one major mechanism of epigenetic regulation. Nuclear antigen Sp100 (SPECKLED, 100 kDa), a constitutive component of the promyelocytic leukemia nuclear bodies, plays key roles in intrinsic immunity and transcriptional repression. Sp100C, a splicing isoform specifically up-regulated upon interferon stimulation, harbors a unique tandem plant homeodomain (PHD) finger and bromodomain at its C terminus. Combining structural, quantitative binding, and cellular co-localization studies, we characterized Sp100C PHD finger as an unmethylated histone H3 Lys(4) (H3K4me0) reader that tolerates histone H3 Thr(3) phosphorylation (H3T3ph), histone H3 Lys(9) trimethylation (H3K9me3), and histone H3 Ser(10) phosphorylation (H3S10ph), hallmarks associated with the mitotic chromosome. In contrast, whereas H3K4me0 reader activity is conserved in Sp140, an Sp100C paralog, the multivalent tolerance of H3T3ph, H3K9me3, and H3S10ph was lost for Sp140. The complex structure determined at 2.1 Å revealed a highly coordinated lysine ϵ-amine recognition sphere formed by an extended N-terminal motif for H3K4me0 readout. Interestingly, reader pocket rigidification by disulfide bond formation enhanced H3K4me0 binding by Sp100C. An additional complex structure solved at 2.7 Å revealed that H3T3ph is recognized by the arginine residue, Arg(713), that is unique to the PHD finger of Sp100C. Consistent with a restrictive cellular role of Sp100C, these results establish a direct chromatin targeting function of Sp100C that may regulate transcriptional gene silencing and promyelocytic leukemia nuclear body-mediated intrinsic immunity in response to interferon stimulation.
- From the Ministry of Education Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084 and.
Organizational Affiliation: