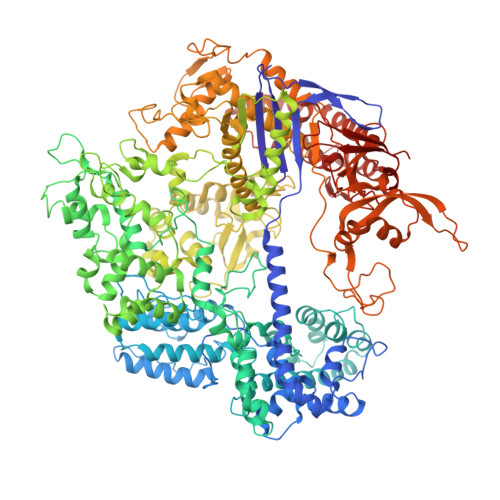
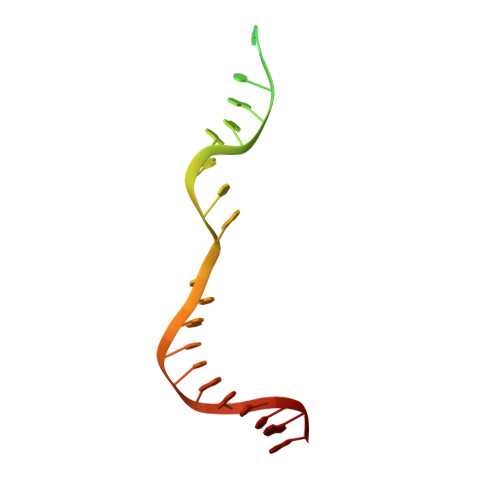
Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage.
Jiang, F., Taylor, D.W., Chen, J.S., Kornfeld, J.E., Zhou, K., Thompson, A.J., Nogales, E., Doudna, J.A.(2016) Science 351: 867-871
- PubMed: 26841432
- DOI: https://doi.org/10.1126/science.aad8282
- Primary Citation of Related Structures:
5F9R - PubMed Abstract:
Bacterial adaptive immunity and genome engineering involving the CRISPR (clustered regularly interspaced short palindromic repeats)-associated (Cas) protein Cas9 begin with RNA-guided DNA unwinding to form an RNA-DNA hybrid and a displaced DNA strand inside the protein. The role of this R-loop structure in positioning each DNA strand for cleavage by the two Cas9 nuclease domains is unknown. We determine molecular structures of the catalytically active Streptococcus pyogenes Cas9 R-loop that show the displaced DNA strand located near the RuvC nuclease domain active site. These protein-DNA interactions, in turn, position the HNH nuclease domain adjacent to the target DNA strand cleavage site in a conformation essential for concerted DNA cutting. Cas9 bends the DNA helix by 30°, providing the structural distortion needed for R-loop formation.
- Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Organizational Affiliation: