Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori.
Moonens, K., Gideonsson, P., Subedi, S., Bugaytsova, J., Romao, E., Mendez, M., Norden, J., Fallah, M., Rakhimova, L., Shevtsova, A., Lahmann, M., Castaldo, G., Brannstrom, K., Coppens, F., Lo, A.W., Ny, T., Solnick, J.V., Vandenbussche, G., Oscarson, S., Hammarstrom, L., Arnqvist, A., Berg, D.E., Muyldermans, S., Boren, T., Remaut, H.(2016) Cell Host Microbe 19: 55-66
- PubMed: 26764597
- DOI: https://doi.org/10.1016/j.chom.2015.12.004
- Primary Citation of Related Structures:
5F7K, 5F7L, 5F7M, 5F7N, 5F7W, 5F7Y, 5F8Q, 5F8R, 5F93, 5F97, 5F9A, 5F9D - PubMed Abstract:
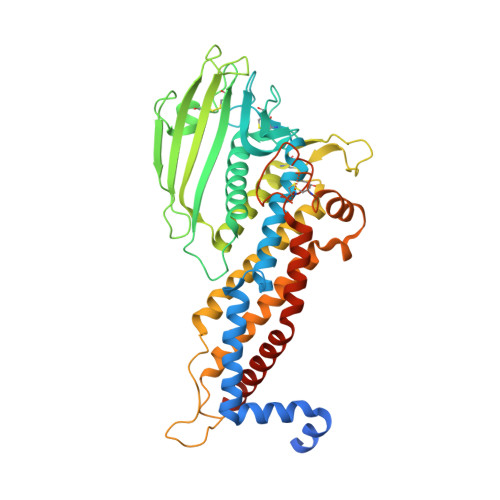
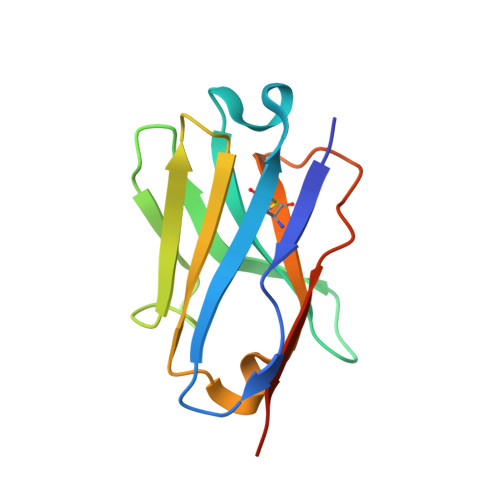
The Helicobacter pylori adhesin BabA binds mucosal ABO/Le(b) blood group (bg) carbohydrates. BabA facilitates bacterial attachment to gastric surfaces, increasing strain virulence and forming a recognized risk factor for peptic ulcers and gastric cancer. High sequence variation causes BabA functional diversity, but the underlying structural-molecular determinants are unknown. We generated X-ray structures of representative BabA isoforms that reveal a polymorphic, three-pronged Le(b) binding site. Two diversity loops, DL1 and DL2, provide adaptive control to binding affinity, notably ABO versus O bg preference. H. pylori strains can switch bg preference with single DL1 amino acid substitutions, and can coexpress functionally divergent BabA isoforms. The anchor point for receptor binding is the embrace of an ABO fucose residue by a disulfide-clasped loop, which is inactivated by reduction. Treatment with the redox-active pharmaceutic N-acetylcysteine lowers gastric mucosal neutrophil infiltration in H. pylori-infected Le(b)-expressing mice, providing perspectives on possible H. pylori eradication therapies.
- Structural and Molecular Microbiology, Structural Biology Research Center, VIB, Pleinlaan 2, 1050 Brussels, Belgium; Structural Biology Brussels, Vrije Universiteit Brussel, Pleinlaan 2, 1050 Brussels, Belgium.
Organizational Affiliation: