FAK Forms a Complex with MEF2 to Couple Biomechanical Signaling to Transcription in Cardiomyocytes.
Cardoso, A.C., Pereira, A.H.M., Ambrosio, A.L.B., Consonni, S.R., Rocha de Oliveira, R., Bajgelman, M.C., Dias, S.M.G., Franchini, K.G.(2016) Structure 24: 1301-1310
- PubMed: 27427476
- DOI: https://doi.org/10.1016/j.str.2016.06.003
- Primary Citation of Related Structures:
5F28 - PubMed Abstract:
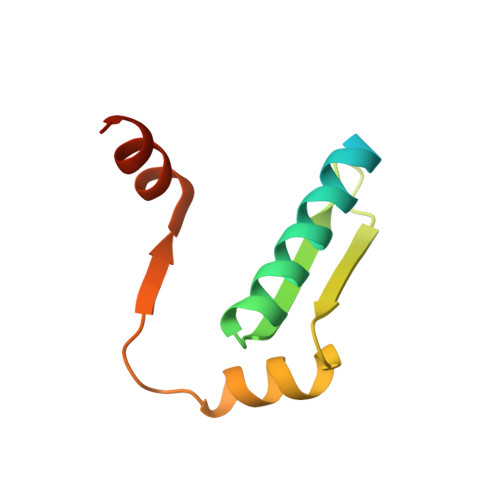
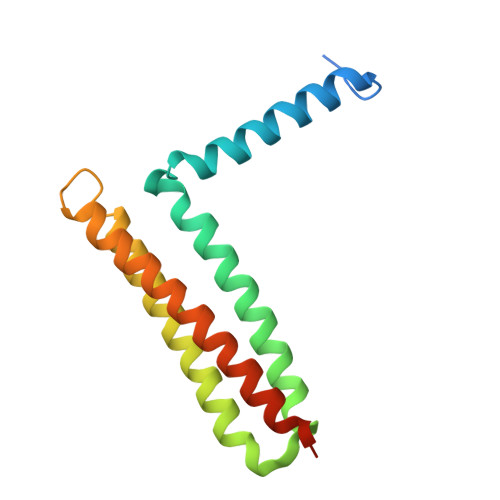
Focal adhesion kinase (FAK) has emerged as a mediator of mechanotransduction in cardiomyocytes, regulating gene expression during hypertrophic remodeling. However, how FAK signaling is relayed onward to the nucleus is unclear. Here, we show that FAK interacts with and regulates myocyte enhancer factor 2 (MEF2), a master cardiac transcriptional regulator. In cardiomyocytes exposed to biomechanical stimulation, FAK accumulates in the nucleus, binds to and upregulates the transcriptional activity of MEF2 through an interaction with the FAK focal adhesion targeting (FAT) domain. In the crystal structure (2.9 Å resolution), FAT binds to a stably folded groove in the MEF2 dimer, known to interact with regulatory cofactors. FAK cooperates with MEF2 to enhance the expression of Jun in cardiomyocytes, an important component of hypertrophic response to mechanical stress. These findings underscore a connection between the mechanotransduction involving FAK and transcriptional regulation by MEF2, with potential relevance to the pathogenesis of cardiac disease.
- Brazilian National Laboratory for Biosciences, Center for Research in Energy and Materials, Campinas, São Paulo 13084-971, Brazil.
Organizational Affiliation: