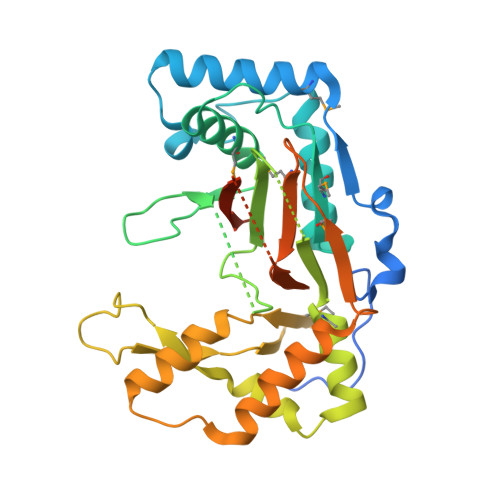
Structural analysis of a phosphonate hydroxylase with an access tunnel at the back of the active site.
Li, C., Junaid, M., Almuqri, E.A., Hao, S., Zhang, H.(2016) Acta Crystallogr F Struct Biol Commun 72: 362-368
- PubMed: 27139827
- DOI: https://doi.org/10.1107/S2053230X16004933
- Primary Citation of Related Structures:
5EQN - PubMed Abstract:
FrbJ is a member of the Fe(2+)/α-ketoglutarate-dependent dioxygenase family which hydroxylates the natural product FR-900098 of Streptomyces rubellomurinus, yielding the phosphonate antibiotic FR-33289. Here, the crystal structure of FrbJ, which shows structural homology to taurine dioxygenase (TauD), a key member of the same family, is reported. Unlike other members of the family, FrbJ has an unusual lid structure which consists of two β-strands with a long loop between them. To investigate the role of this lid motif, a molecular-dynamics simulation was performed with the FrbJ structure. The molecular-dynamics simulation analysis implies that the lid-loop region is highly flexible, which is consistent with the fact that FrbJ has a relatively broad spectrum of substrates with different lengths. Interestingly, an access tunnel is found at the back of the active site which connects the putative binding site of α-ketoglutarate to the solvent outside.
- Department of Biotechnology, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, People's Republic of China.
Organizational Affiliation: