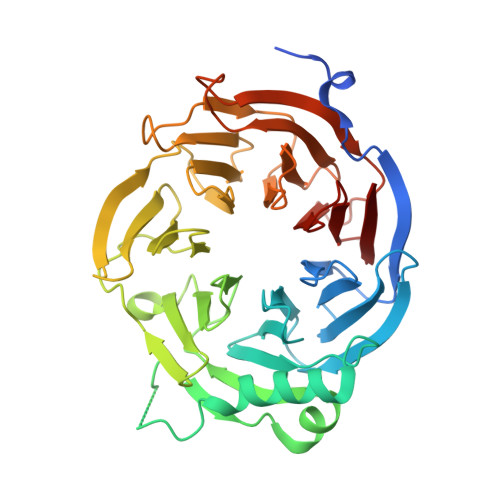
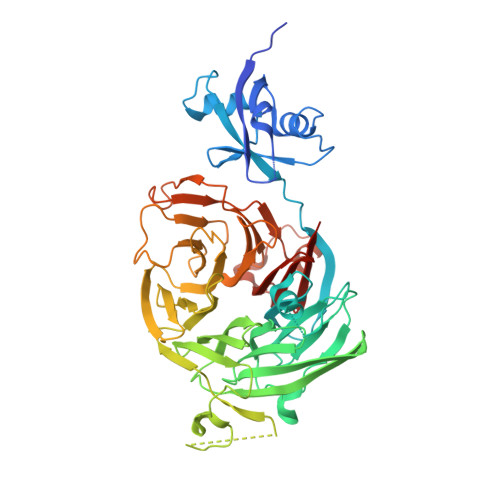
Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface.
Thoms, M., Ahmed, Y.L., Maddi, K., Hurt, E., Sinning, I.(2016) Nucleic Acids Res 44: 926-939
- PubMed: 26657628
- DOI: https://doi.org/10.1093/nar/gkv1365
- Primary Citation of Related Structures:
5EM2 - PubMed Abstract:
The complicated process of eukaryotic ribosome biogenesis involves about 200 assembly factors that transiently associate with the nascent pre-ribosome in a spatiotemporally ordered way. During the early steps of 60S subunit formation, several proteins, collectively called A3 cluster factors, participate in the removal of the internal transcribed spacer 1 (ITS1) from 27SA3 pre-rRNA. Among these factors is the conserved hetero-trimeric Nop7-Erb1-Ytm1 complex (or human Pes1-Bop1-Wdr12), which is removed from the evolving pre-60S particle by the AAA ATPase Rea1 to allow progression in the pathway. Here, we clarify how Ytm1 and Erb1 interact, which has implications for the release mechanism of both factors from the pre-ribosome. Biochemical studies show that Ytm1 and Erb1 bind each other via their ß-propeller domains. The crystal structure of the Erb1-Ytm1 heterodimer determined at 2.67Å resolution reveals an extended interaction surface between the propellers in a rarely observed binding mode. Structure-based mutations in the interface that impair the Erb1-Ytm1 interaction do not support growth, with specific defects in 60S subunit synthesis. Under these mutant conditions, it becomes clear that an intact Erb1-Ytm1 complex is required for 60S maturation and that loss of this stable interaction prevents ribosome production.
- Heidelberg University Biochemistry Center (BZH), INF 328, D-69120 Heidelberg, Germany.
Organizational Affiliation: