Structural basis of nonribosomal peptide macrocyclization in fungi
Zhang, J., Liu, N., Cacho, R.A., Gong, Z., Liu, Z., Qin, W., Tang, C., Tang, Y., Zhou, J.(2016) Nat Chem Biol 12: 1001-1003
- PubMed: 27748753
- DOI: https://doi.org/10.1038/nchembio.2202
- Primary Citation of Related Structures:
5DIJ, 5DLK, 5EGF, 5EJD - PubMed Abstract:
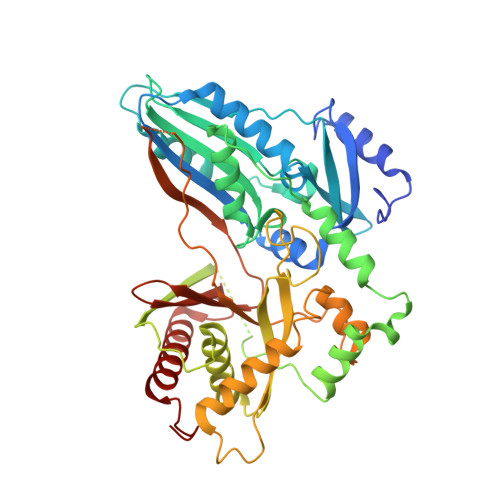
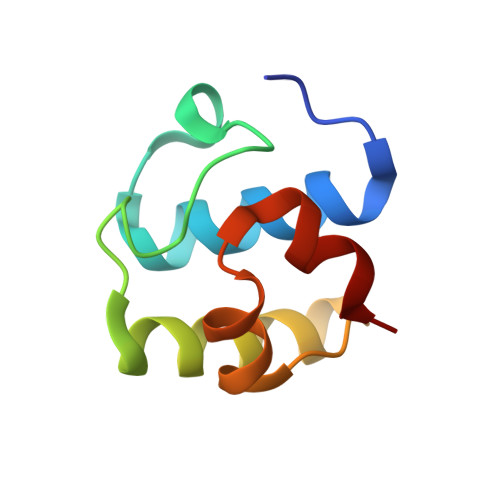
Nonribosomal peptide synthetases (NRPSs) in fungi biosynthesize important pharmaceutical compounds, including penicillin, cyclosporine and echinocandin. To understand the fungal strategy of forging the macrocyclic peptide linkage, we determined the crystal structures of the terminal condensation-like (C T ) domain and the holo thiolation (T)-C T complex of Penicillium aethiopicum TqaA. The first, to our knowledge, structural depiction of the terminal module in a fungal NRPS provides a molecular blueprint for generating new macrocyclic peptide natural products.
- State Key Laboratory of Bio-organic and Natural Products Chemistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, Shanghai 200032, China.
Organizational Affiliation: