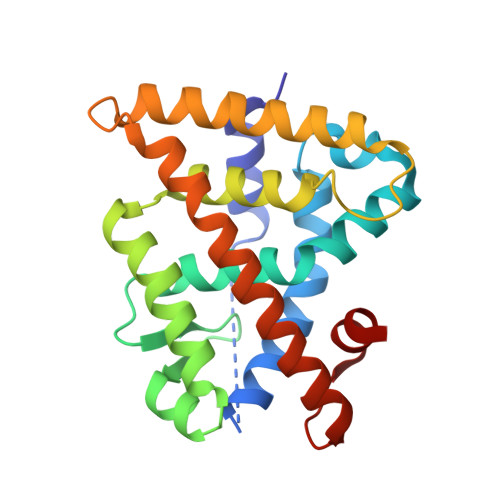
Chiral Dihydrobenzofuran Acids Show Potent Retinoid X Receptor-Nuclear Receptor Related 1 Protein Dimer Activation.
Sunden, H., Schafer, A., Scheepstra, M., Leysen, S., Malo, M., Ma, J.N., Burstein, E.S., Ottmann, C., Brunsveld, L., Olsson, R.(2016) J Med Chem 59: 1232-1238
- PubMed: 26820900
- DOI: https://doi.org/10.1021/acs.jmedchem.5b01702
- Primary Citation of Related Structures:
5EC9 - PubMed Abstract:
The nuclear receptor Nurr1 can be activated by RXR via heterodimerization (RXR-Nurr1) and is a promising target for treating neurodegenerative diseases. We herein report the enantioselective synthesis and SAR of sterically constricted benzofurans at RXR. The established SAR, using whole cell functional assays, lead to the full agonist 9a at RXR (pEC50 of 8.2) and RXR-Nurr1. The X-ray structure shows enantiomeric discrimination where 9a optimally addresses the ligand binding pocket of RXR.
- Department of Chemistry and Molecular Biology, Medicinal Chemistry, University of Gothenburg , SE-41296 Gothenburg, Sweden.
Organizational Affiliation: