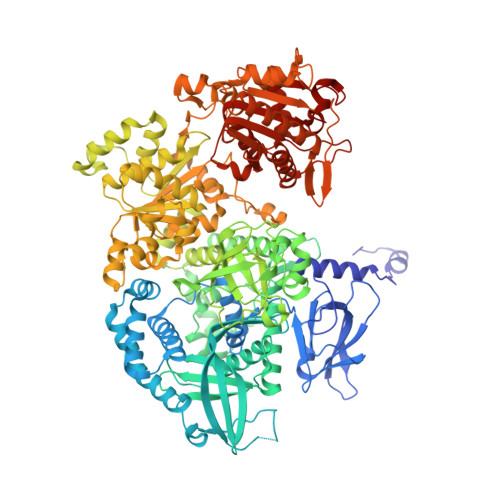
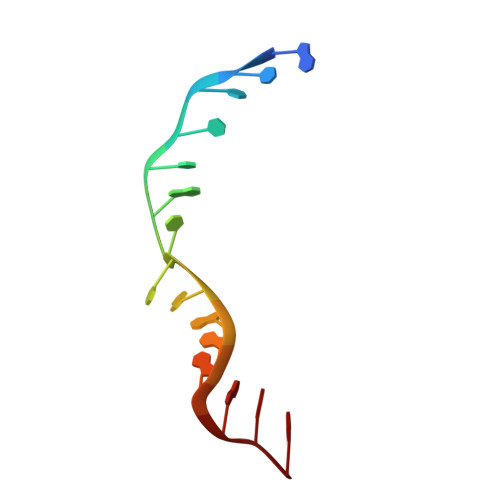
Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Zhou, C., Pourmal, S., Pavletich, N.P.(2015) Elife 4
- PubMed: 26491943
- DOI: https://doi.org/10.7554/eLife.09832
- Primary Citation of Related Structures:
5EAN, 5EAW, 5EAX, 5EAY - PubMed Abstract:
The Dna2 nuclease-helicase maintains genomic integrity by processing DNA double-strand breaks, Okazaki fragments and stalled replication forks. Dna2 requires ssDNA ends, and is dependent on the ssDNA-binding protein Rpa, which controls cleavage polarity. Here we present the 2.3 Å structure of intact mouse Dna2 bound to a 15-nucleotide ssDNA. The nuclease active site is embedded in a long, narrow tunnel through which the DNA has to thread. The helicase domain is required for DNA binding but not threading. We also present the structure of a flexibly-tethered Dna2-Rpa interaction that recruits Dna2 to Rpa-coated DNA. We establish that a second Dna2-Rpa interaction is mutually exclusive with Rpa-DNA interactions and mediates the displacement of Rpa from ssDNA. This interaction occurs at the nuclease tunnel entrance and the 5' end of the Rpa-DNA complex. Hence, it only displaces Rpa from the 5' but not 3' end, explaining how Rpa regulates cleavage polarity.
- Structural Biology Program, Memorial Sloan Kettering Cancer Center, New York, United States.
Organizational Affiliation: