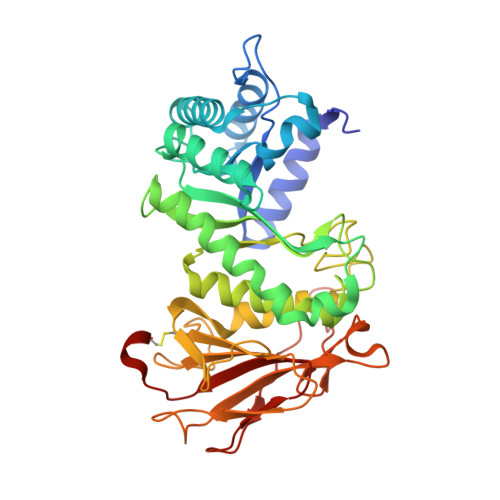
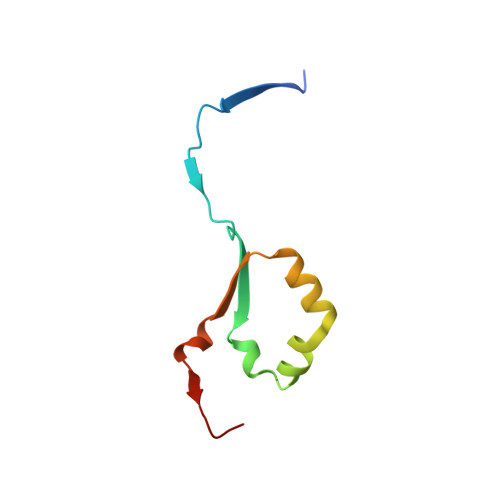
Structural characterization of human heparanase reveals insights into substrate recognition.
Wu, L., Viola, C.M., Brzozowski, A.M., Davies, G.J.(2015) Nat Struct Mol Biol 22: 1016-1022
- PubMed: 26575439
- DOI: https://doi.org/10.1038/nsmb.3136
- Primary Citation of Related Structures:
5E8M, 5E97, 5E98, 5E9B, 5E9C - PubMed Abstract:
Heparan sulfate (HS) is a glycosaminoglycan that forms a key component of the extracellular matrix (ECM). Breakdown of HS is carried out by heparanase (HPSE), an endo-β-glucuronidase of the glycoside hydrolase 79 (GH79) family. Overexpression of HPSE results in breakdown of extracellular HS and release of stored growth factors and hence is strongly linked to cancer metastasis. Here we present crystal structures of human HPSE at 1.6-Å to 1.9-Å resolution that reveal how an endo-acting binding cleft is exposed by proteolytic activation of latent proHPSE. We used oligosaccharide complexes to map the substrate-binding and sulfate-recognition motifs. These data shed light on the structure and interactions of a key enzyme involved in ECM maintenance and provide a starting point for the design of HPSE inhibitors for use as biochemical tools and anticancer therapeutics.
- Department of Chemistry, University of York, York, UK.
Organizational Affiliation: