High resolution crystal structure of LuxS - Quorum sensor molecular complex from Salmonella typhi at 1.58 Angstroms
Perumal, P., Raina, R., Arockiasamy, A., SundaraBaalaji, N.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| S-ribosylhomocysteine lyase | 179 | Salmonella enterica subsp. enterica serovar Typhi | Mutation(s): 1 Gene Names: luxS, STY2943, t2714 EC: 4.4.1.21 | 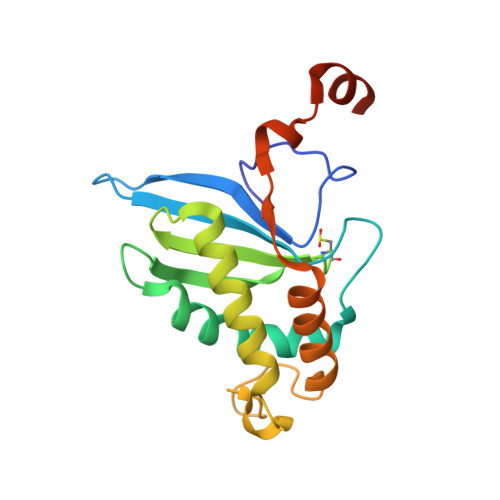 | |
UniProt | |||||
Find proteins for Q8Z4D7 (Salmonella typhi) Explore Q8Z4D7 Go to UniProtKB: Q8Z4D7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8Z4D7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PAV Query on PAV | D [auth A] | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran C5 H10 O5 BVIYGXUQVXBHQS-IUYQGCFVSA-N |  | ||
| MET Query on MET | F [auth B] | METHIONINE C5 H11 N O2 S FFEARJCKVFRZRR-BYPYZUCNSA-N |  | ||
| ZN Query on ZN | C [auth A], E [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSD Query on CSD | A, B | L-PEPTIDE LINKING | C3 H7 N O4 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 48.345 | α = 90 |
| b = 66.427 | β = 106.17 |
| c = 54.31 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data processing |
| BALBES | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Indian Council of Medical Research | India | 2011-15260 |