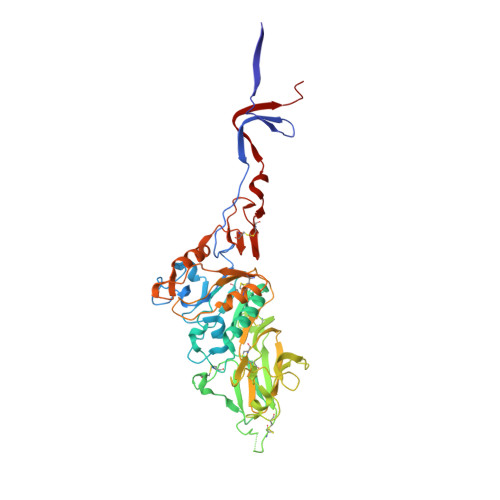
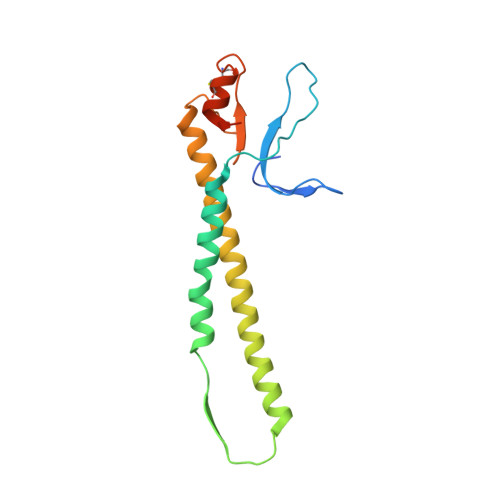
An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
Song, H., Qi, J., Khedri, Z., Diaz, S., Yu, H., Chen, X., Varki, A., Shi, Y., Gao, G.F.(2016) PLoS Pathog 12: e1005411-e1005411
- PubMed: 26816272
- DOI: https://doi.org/10.1371/journal.ppat.1005411
- Primary Citation of Related Structures:
5E5W, 5E62, 5E64, 5E65, 5E66 - PubMed Abstract:
Influenza viruses cause seasonal flu each year and pandemics or epidemic sporadically, posing a major threat to public health. Recently, a new influenza D virus (IDV) was isolated from pigs and cattle. Here, we reveal that the IDV utilizes 9-O-acetylated sialic acids as its receptor for virus entry. Then, we determined the crystal structures of hemagglutinin-esterase-fusion glycoprotein (HEF) of IDV both in its free form and in complex with the receptor and enzymatic substrate analogs. The IDV HEF shows an extremely similar structural fold as the human-infecting influenza C virus (ICV) HEF. However, IDV HEF has an open receptor-binding cavity to accommodate diverse extended glycan moieties. This structural difference provides an explanation for the phenomenon that the IDV has a broad cell tropism. As IDV HEF is structurally and functionally similar to ICV HEF, our findings highlight the potential threat of the virus to public health.
- CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: