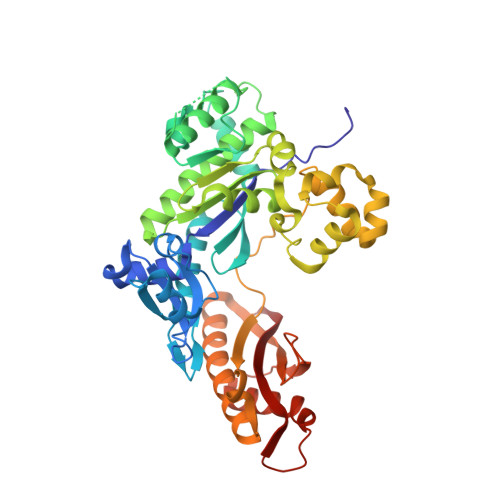
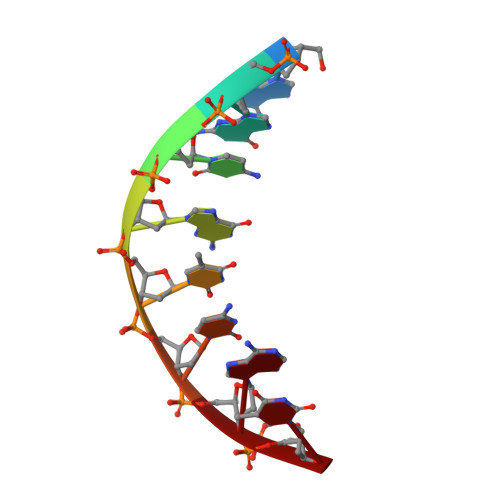
Lesion Orientation ofO4-Alkylthymidine Influences Replication by Human DNA Polymeraseeta.
O'Flaherty, D.K., Patra, A., Su, Y., Guengerich, F.P., Egli, M., Wilds, C.J.(2016) Chem Sci 7: 4896-4904
- PubMed: 27574558
- DOI: https://doi.org/10.1039/C6SC00666C
- Primary Citation of Related Structures:
5DLF, 5DLG, 5DQG, 5DQH, 5DQI - PubMed Abstract:
DNA lesions that elude repair may undergo translesion synthesis catalyzed by Y-family DNA polymerases. O 4 -Alkylthymidines, persistent adducts that can result from carcinogenic agents, may be encountered by DNA polymerases. The influence of lesion orientation around the C4- O 4 bond on processing by human DNA polymerase η (hPol η ) was studied for oligonucleotides containing O 4 -methylthymidine, O 4 -ethylthymidine, and analogs restricting the O 4 -methylene group in an anti -orientation. Primer extension assays revealed that the O 4 -alkyl orientation influences hPol η bypass. Crystal structures of hPol η •DNA•dNTP ternary complexes with O 4 -methyl- or O 4 -ethylthymidine in the template strand showed the nucleobase of the former lodged near the ceiling of the active site, with the syn - O 4 -methyl group engaged in extensive hydrophobic interactions. This unique arrangement for O 4 -methylthymidine with hPol η , inaccessible for the other analogs due to steric/conformational restriction, is consistent with differences observed for nucleotide incorporation and supports the concept that lesion conformation influences extension across DNA damage. Together, these results provide mechanistic insights on the mutagenicity of O 4 MedT and O 4 EtdT when acted upon by hPol η .
- Department of Chemistry and Biochemistry, Concordia University, Montréal, Québec H4B1R6, Canada.
Organizational Affiliation: