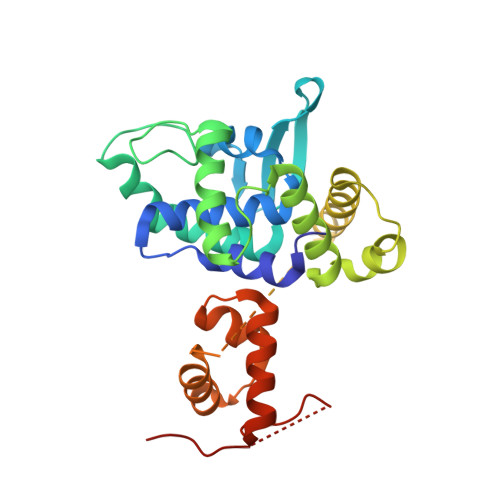
Activation of Xer-recombination at dif: structural basis of the FtsK gamma-XerD interaction.
Keller, A.N., Xin, Y., Boer, S., Reinhardt, J., Baker, R., Arciszewska, L.K., Lewis, P.J., Sherratt, D.J., Lowe, J., Grainge, I.(2016) Sci Rep 6: 33357-33357
- PubMed: 27708355
- DOI: https://doi.org/10.1038/srep33357
- Primary Citation of Related Structures:
5DCF - PubMed Abstract:
Bacterial chromosomes are most often circular DNA molecules. This can produce a topological problem; a genetic crossover from homologous recombination results in dimerization of the chromosome. A chromosome dimer is lethal unless resolved. A site-specific recombination system catalyses this dimer-resolution reaction at the chromosomal site dif. In Escherichia coli, two tyrosine-family recombinases, XerC and XerD, bind to dif and carry out two pairs of sequential strand exchange reactions. However, what makes the reaction unique among site-specific recombination reactions is that the first step, XerD-mediated strand exchange, relies on interaction with the very C-terminus of the FtsK DNA translocase. FtsK is a powerful molecular motor that functions in cell division, co-ordinating division with clearing chromosomal DNA from the site of septation and also acts to position the dif sites for recombination. This is a model system for unlinking, separating and segregating large DNA molecules. Here we describe the molecular detail of the interaction between XerD and FtsK that leads to activation of recombination as deduced from a co-crystal structure, biochemical and in vivo experiments. FtsKγ interacts with the C-terminal domain of XerD, above a cleft where XerC is thought to bind. We present a model for activation of recombination based on structural data.
- School of Environmental and Life Sciences, University of Newcastle, University Drive, Callaghan NSW 2308, Australia.
Organizational Affiliation: