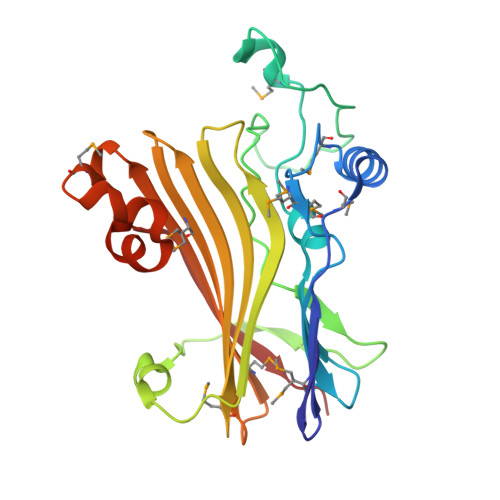
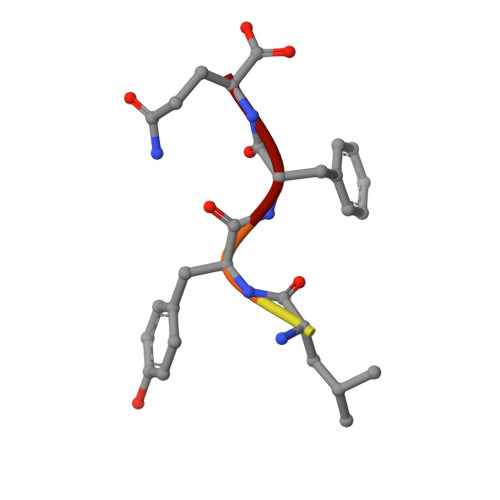
Crystal structure of the human Tip41 orthologue, TIPRL, reveals a novel fold and a binding site for the PP2Ac C-terminus.
Scorsato, V., Lima, T.B., Righetto, G.L., Zanchin, N.I., Brandao-Neto, J., Sandy, J., Pereira, H.D., Ferrari, A.J., Gozzo, F.C., Smetana, J.H., Aparicio, R.(2016) Sci Rep 6: 30813-30813
- PubMed: 27489114
- DOI: https://doi.org/10.1038/srep30813
- Primary Citation of Related Structures:
5D9G - PubMed Abstract:
TOR signaling pathway regulator-like (TIPRL) is a regulatory protein which inhibits the catalytic subunits of Type 2A phosphatases. Several cellular contexts have been proposed for TIPRL, such as regulation of mTOR signaling, inhibition of apoptosis and biogenesis and recycling of PP2A, however, the underlying molecular mechanism is still poorly understood. We have solved the crystal structure of human TIPRL at 2.15 Å resolution. The structure is a novel fold organized around a central core of antiparallel beta-sheet, showing an N-terminal α/β region at one of its surfaces and a conserved cleft at the opposite surface. Inside this cleft, we found a peptide derived from TEV-mediated cleavage of the affinity tag. We show by mutagenesis, pulldown and hydrogen/deuterium exchange mass spectrometry that this peptide is a mimic for the conserved C-terminal tail of PP2A, an important region of the phosphatase which regulates holoenzyme assembly, and TIPRL preferentially binds the unmodified version of the PP2A-tail mimetic peptide DYFL compared to its tyrosine-phosphorylated version. A docking model of the TIPRL-PP2Ac complex suggests that TIPRL blocks the phosphatase's active site, providing a structural framework for the function of TIPRL in PP2A inhibition.
- Institute of Chemistry, University of Campinas, Campinas, São Paulo 13083-970, Brazil.
Organizational Affiliation: