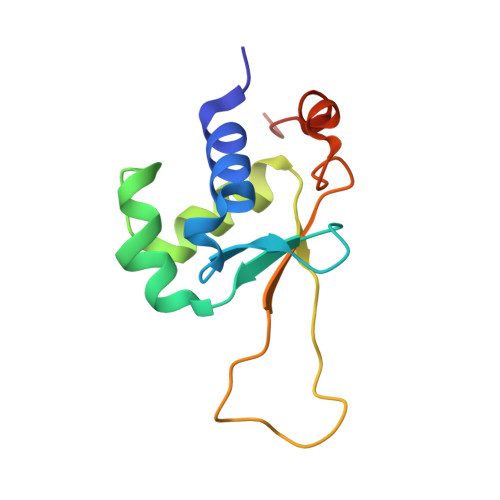
Structures of HSF2 reveal mechanisms for differential regulation of human heat-shock factors.
Jaeger, A.M., Pemble, C.W., Sistonen, L., Thiele, D.J.(2016) Nat Struct Mol Biol 23: 147-154
- PubMed: 26727490
- DOI: https://doi.org/10.1038/nsmb.3150
- Primary Citation of Related Structures:
5D8K, 5D8L - PubMed Abstract:
Heat-shock transcription factor (HSF) family members function in stress protection and in human diseases including proteopathies, neurodegeneration and cancer. The mechanisms that drive distinct post-translational modifications, cofactor recruitment and target-gene activation for specific HSF paralogs are unknown. We present crystal structures of the human HSF2 DNA-binding domain (DBD) bound to DNA, revealing an unprecedented view of HSFs that provides insights into their unique biology. The HSF2 DBD structures resolve a new C-terminal helix that directs wrapping of the coiled-coil domain around DNA, thereby exposing paralog-specific sequences of the DBD surface for differential post-translational modifications and cofactor interactions. We further demonstrate a direct interaction between HSF1 and HSF2 through their coiled-coil domains. Together, these features provide a new model for HSF structure as the basis for differential and combinatorial regulation, which influences the transcriptional response to cellular stress.
- Department of Pharmacology and Cancer Biology, Duke University School of Medicine, Durham, North Carolina, USA.
Organizational Affiliation: