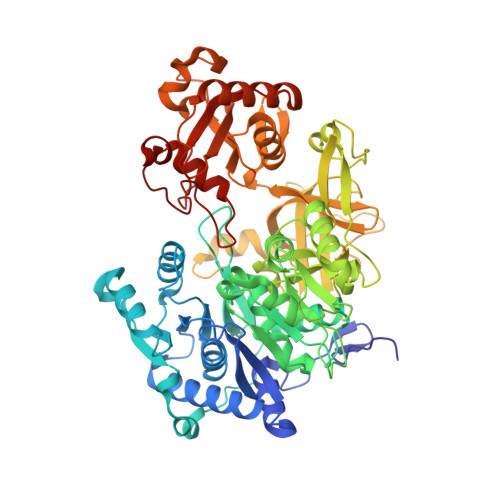
Crystal structure of FadD32, an enzyme essential for mycolic acid biosynthesis in mycobacteria.
Li, W., Gu, S., Fleming, J., Bi, L.(2015) Sci Rep 5: 15493-15493
- PubMed: 26628098
- DOI: https://doi.org/10.1038/srep15493
- Primary Citation of Related Structures:
5D6J, 5D6N - PubMed Abstract:
Fatty acid degradation protein D32 (FadD32), an enzyme required for mycolic acid biosynthesis and essential for mycobacterial growth, has recently been identified as a valid and promising target for anti-tuberculosis drug development. Here we report the crystal structures of Mycobacterium smegmatis FadD32 in the apo and ATP-bound states at 2.4 Å and 2.25 Å resolution, respectively. FadD32 consists of two globular domains connected by a flexible linker. ATP binds in a cleft at the interface between the N- and C-terminal domains and its binding induces significant local conformational changes in FadD32. The binding sites of meromycolic acid and phosphopantetheine are identified by structural comparison with other members of the adenylating enzyme superfamily. These results will improve our understanding of the catalytic mechanism of FadD32 and help in the design of inhibitors of this essential enzyme.
- Key Laboratory of RNA Biology, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: