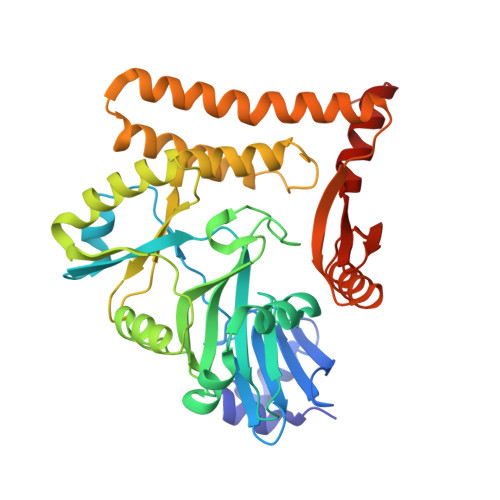
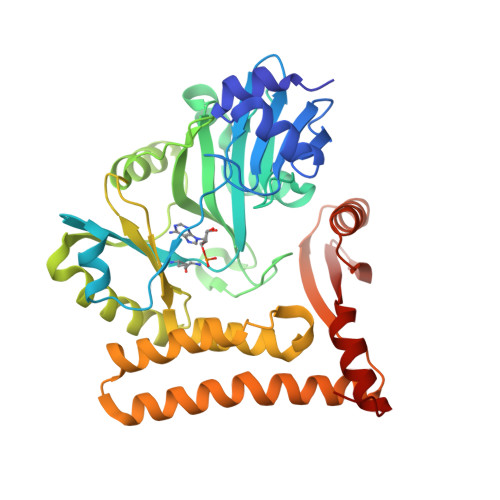
Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis.
Gu, H., Yoshinari, S., Ghosh, R., Ignatochkina, A.V., Gollnick, P.D., Murakami, K.S., Ho, C.K.(2016) Nucleic Acids Res 44: 2337-2347
- PubMed: 26896806
- DOI: https://doi.org/10.1093/nar/gkw094
- Primary Citation of Related Structures:
5D1O, 5D1P - PubMed Abstract:
An ATP-dependent RNA ligase from Methanobacterium thermoautotrophicum (MthRnl) catalyzes intramolecular ligation of single-stranded RNA to form a closed circular RNA via covalent ligase-AMP and RNA-adenylylate intermediate. Here, we report the X-ray crystal structures of an MthRnl•ATP complex as well as the covalent MthRnl-AMP intermediate. We also performed structure-guided mutational analysis to survey the functions of 36 residues in three component steps of the ligation pathway including ligase-adenylylation (step 1), RNA adenylylation (step 2) and phosphodiester bond synthesis (step 3). Kinetic analysis underscored the importance of motif 1a loop structure in promoting phosphodiester bond synthesis. Alanine substitutions of Thr117 or Arg118 favor the reverse step 2 reaction to deadenylate the 5'-AMP from the RNA-adenylate, thereby inhibiting step 3 reaction. Tyr159, Phe281 and Glu285, which are conserved among archaeal ATP-dependent RNA ligases and are situated on the surface of the enzyme, are required for RNA binding. We propose an RNA binding interface of the MthRnl based on the mutational studies and two sulfate ions that co-crystallized at the active site cleft in the MthRnl-AMP complex.
- Department of Biological Sciences, State University of New York, Buffalo, NY 14260, USA.
Organizational Affiliation: