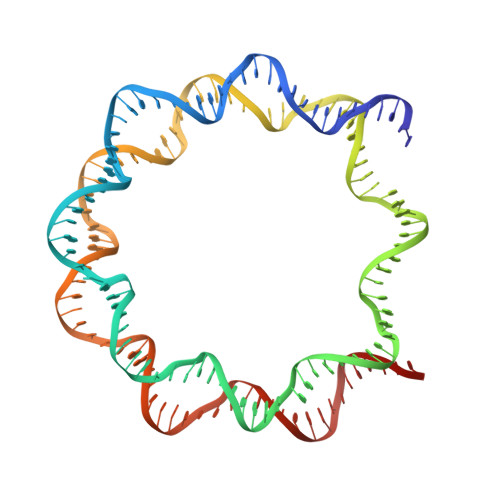
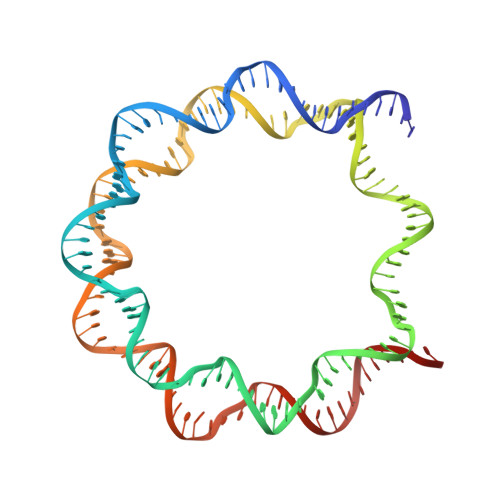
An Organometallic Compound which Exhibits a DNA Topology-Dependent One-Stranded Intercalation Mode.
Ma, Z., Palermo, G., Adhireksan, Z., Murray, B.S., von Erlach, T., Dyson, P.J., Rothlisberger, U., Davey, C.A.(2016) Angew Chem Int Ed Engl 55: 7441-7444
- PubMed: 27184539
- DOI: https://doi.org/10.1002/anie.201602145
- Primary Citation of Related Structures:
5CP6 - PubMed Abstract:
Understanding how small molecules interact with DNA is essential since it underlies a multitude of pathological conditions and therapeutic interventions. Many different intercalator compounds have been studied because of their activity as mutagens or drugs, but little is known regarding their interaction with nucleosomes, the protein-packaged form of DNA in cells. Here, using crystallographic methods and molecular dynamics simulations, we discovered that adducts formed by [(η(6) -THA)Ru(ethylenediamine)Cl][PF6 ] (THA=5,8,9,10-tetrahydroanthracene; RAED-THA-Cl[PF6 ]) in the nucleosome comprise a novel one-stranded intercalation and DNA distortion mode. Conversely, the THA group in fact remains solvent exposed and does not disrupt base stacking in RAED-THA adducts on B-form DNA. This newly observed DNA binding mode and topology dependence may actually be prevalent and should be considered when studying covalently binding intercalating compounds.
- School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore, 637551, Singapore.
Organizational Affiliation: