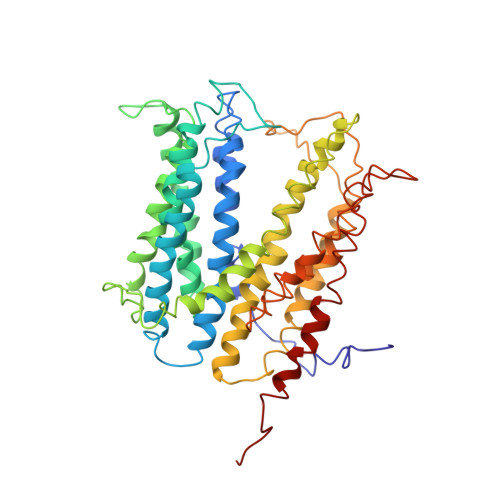
Structural basis for the blockade of MATE multidrug efflux pumps.
Radchenko, M., Symersky, J., Nie, R., Lu, M.(2015) Nat Commun 6: 7995-7995
- PubMed: 26246409
- DOI: https://doi.org/10.1038/ncomms8995
- Primary Citation of Related Structures:
5C6N, 5C6O, 5C6P - PubMed Abstract:
Multidrug and toxic compound extrusion (MATE) transporters underpin multidrug resistance by using the H(+) or Na(+) electrochemical gradient to extrude different drugs across cell membranes. MATE transporters can be further parsed into the DinF, NorM and eukaryotic subfamilies based on their amino-acid sequence similarity. Here we report the 3.0 Å resolution X-ray structures of a protonation-mimetic mutant of an H(+)-coupled DinF transporter, as well as of an H(+)-coupled DinF and a Na(+)-coupled NorM transporters in complexes with verapamil, a small-molecule pharmaceutical that inhibits MATE-mediated multidrug extrusion. Combining structure-inspired mutational and functional studies, we confirm the biological relevance of our crystal structures, reveal the mechanistic differences among MATE transporters, and suggest how verapamil inhibits MATE-mediated multidrug efflux. Our findings offer insights into how MATE transporters extrude chemically and structurally dissimilar drugs and could inform the design of new strategies for tackling multidrug resistance.
- Department of Biochemistry and Molecular Biology, Rosalind Franklin University of Medicine and Science, 3333 Green Bay Road, North Chicago, Illinois 60064, USA.
Organizational Affiliation: