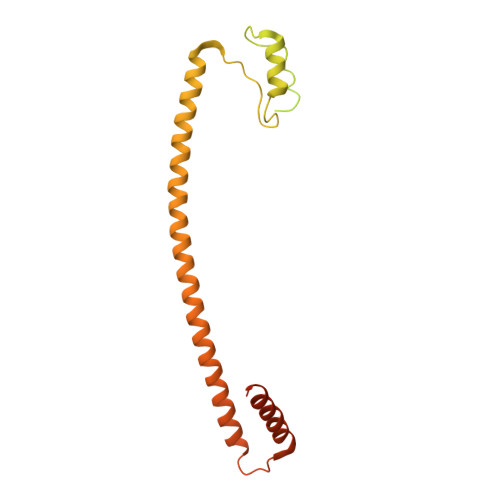
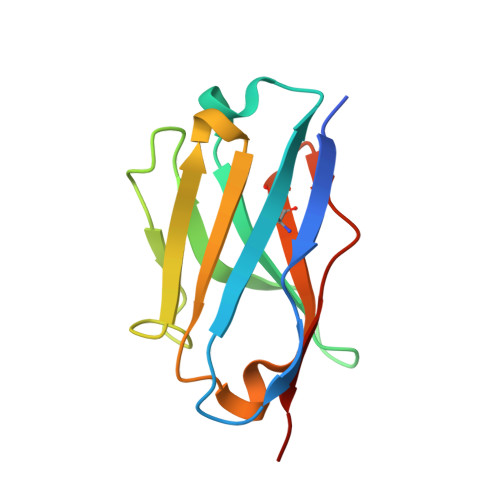
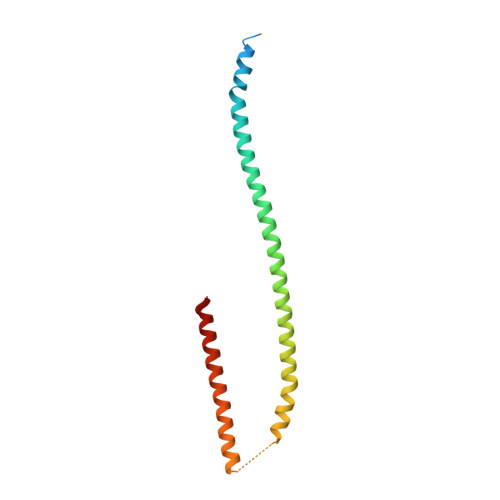Crystal structure of the metazoan Nup62Nup58Nup54 nucleoporin complex.
Chug, H., Trakhanov, S., Hulsmann, B.B., Pleiner, T., Gorlich, D.(2015) Science 350: 106-110
- PubMed: 26292704
- DOI: https://doi.org/10.1126/science.aac7420
- Primary Citation of Related Structures:
5C2U, 5C3L - PubMed Abstract:
Nuclear pore complexes (NPCs) conduct nucleocytoplasmic transport and gain transport selectivity through nucleoporin FG domains. Here, we report a structural analysis of the FG Nup62•58•54 complex, which is a crucial component of the transport system. It comprises a ≈13 nanometer-long trimerization interface with an unusual 2W3F coil, a canonical heterotrimeric coiled coil, and a kink that enforces a compact six-helix bundle. Nup54 also contains a ferredoxin-like domain. We further identified a heterotrimeric Nup93-binding module for NPC anchorage. The quaternary structure alternations in the Nup62 complex, which were previously proposed to trigger a general gating of the NPC, are incompatible with the trimer structure. We suggest that the highly elongated Nup62 complex projects barrier-forming FG repeats far into the central NPC channel, supporting a barrier that guards the entire cross section.
- Department of Cellular Logistics, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany.
Organizational Affiliation: