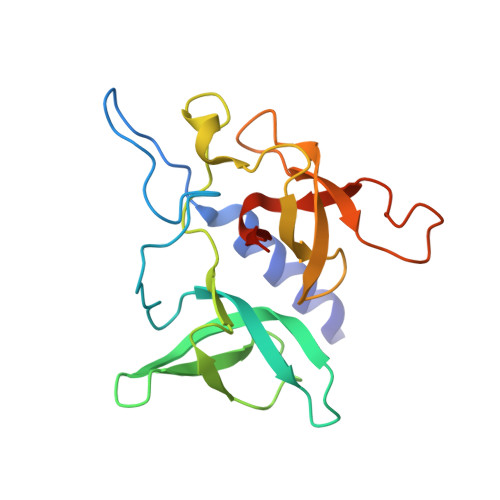
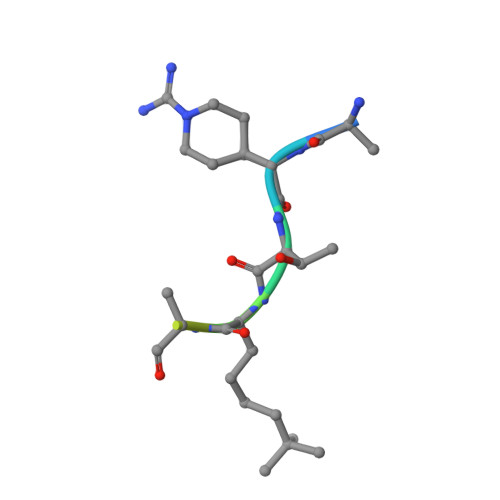
Chemical basis for the recognition of trimethyllysine by epigenetic reader proteins.
Kamps, J.J., Huang, J., Poater, J., Xu, C., Pieters, B.J., Dong, A., Min, J., Sherman, W., Beuming, T., Matthias Bickelhaupt, F., Li, H., Mecinovic, J.(2015) Nat Commun 6: 8911-8911
- PubMed: 26578293
- DOI: https://doi.org/10.1038/ncomms9911
- Primary Citation of Related Structures:
5C0M - PubMed Abstract:
A large number of structurally diverse epigenetic reader proteins specifically recognize methylated lysine residues on histone proteins. Here we describe comparative thermodynamic, structural and computational studies on recognition of the positively charged natural trimethyllysine and its neutral analogues by reader proteins. This work provides experimental and theoretical evidence that reader proteins predominantly recognize trimethyllysine via a combination of favourable cation-π interactions and the release of the high-energy water molecules that occupy the aromatic cage of reader proteins on the association with the trimethyllysine side chain. These results have implications in rational drug design by specifically targeting the aromatic cage of readers of trimethyllysine.
- Institute for Molecules and Materials, Radboud University, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands.
Organizational Affiliation: