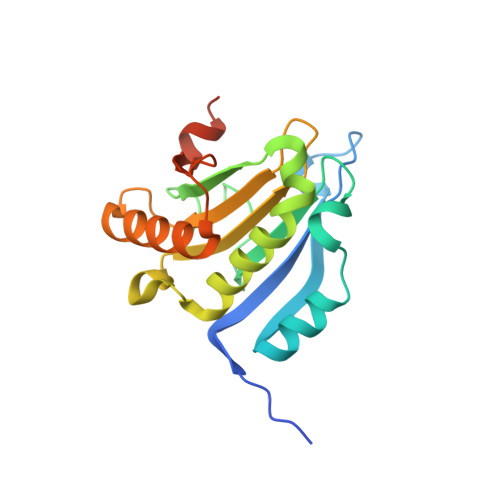
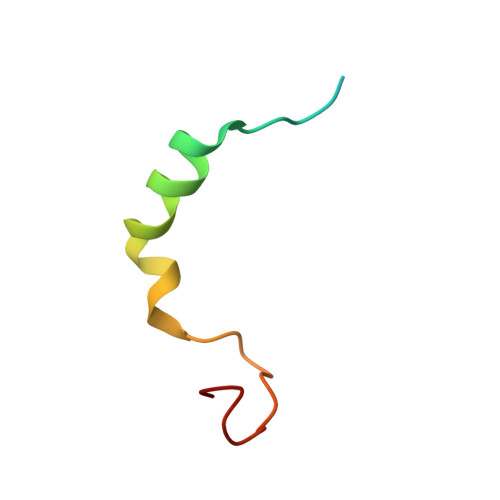
Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1.
Sekiyama, N., Arthanari, H., Papadopoulos, E., Rodriguez-Mias, R.A., Wagner, G., Leger-Abraham, M.(2015) Proc Natl Acad Sci U S A 112: E4036-E4045
- PubMed: 26170285
- DOI: https://doi.org/10.1073/pnas.1512118112
- Primary Citation of Related Structures:
5BXV - PubMed Abstract:
The eIF4E-binding protein (4E-BP) is a phosphorylation-dependent regulator of protein synthesis. The nonphosphorylated or minimally phosphorylated form binds translation initiation factor 4E (eIF4E), preventing binding of eIF4G and the recruitment of the small ribosomal subunit. Signaling events stimulate serial phosphorylation of 4E-BP, primarily by mammalian target of rapamycin complex 1 (mTORC1) at residues T37/T46, followed by T70 and S65. Hyperphosphorylated 4E-BP dissociates from eIF4E, allowing eIF4E to interact with eIF4G and translation initiation to resume. Because overexpression of eIF4E is linked to cellular transformation, 4E-BP is a tumor suppressor, and up-regulation of its activity is a goal of interest for cancer therapy. A recently discovered small molecule, eIF4E/eIF4G interaction inhibitor 1 (4EGI-1), disrupts the eIF4E/eIF4G interaction and promotes binding of 4E-BP1 to eIF4E. Structures of 14- to 16-residue 4E-BP fragments bound to eIF4E contain the eIF4E consensus binding motif, (54)YXXXXLΦ(60) (motif 1) but lack known phosphorylation sites. We report here a 2.1-Å crystal structure of mouse eIF4E in complex with m(7)GTP and with a fragment of human 4E-BP1, extended C-terminally from the consensus-binding motif (4E-BP150-84). The extension, which includes a proline-turn-helix segment (motif 2) followed by a loop of irregular structure, reveals the location of two phosphorylation sites (S65 and T70). Our major finding is that the C-terminal extension (motif 3) is critical to 4E-BP1-mediated cell cycle arrest and that it partially overlaps with the binding site of 4EGI-1. The binding of 4E-BP1 and 4EGI-1 to eIF4E is therefore not mutually exclusive, and both ligands contribute to shift the equilibrium toward the inhibition of translation initiation.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115.
Organizational Affiliation: