Highly dynamic nature of ERdj5 is essential for enhancement of the ER associated degradation
Maegawa, K., Watanabe, S., Okumura, M., Noi, K., Inoue, M., Ushioda, R., Ogura, T., Nagata, K., Inaba, K.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DnaJ homolog subfamily C member 10 | 783 | Mus musculus | Mutation(s): 10 Gene Names: Dnajc10, Erdj5, Jpdi EC: 1.8.4 (PDB Primary Data), 1.8.4.2 (UniProt) | 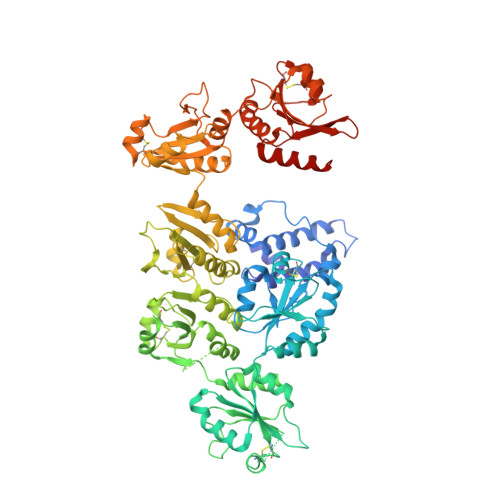 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9DC23 (Mus musculus) Explore Q9DC23 Go to UniProtKB: Q9DC23 | |||||
IMPC: MGI:1914111 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9DC23 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 1PS Query on 1PS | B [auth A] | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE C8 H11 N O3 S REEBJQTUIJTGAL-UHFFFAOYSA-N |  | ||
| CL Query on CL | C [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 91.99 | α = 90 |
| b = 52.97 | β = 113.54 |
| c = 92.9 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| MOLREP | phasing |