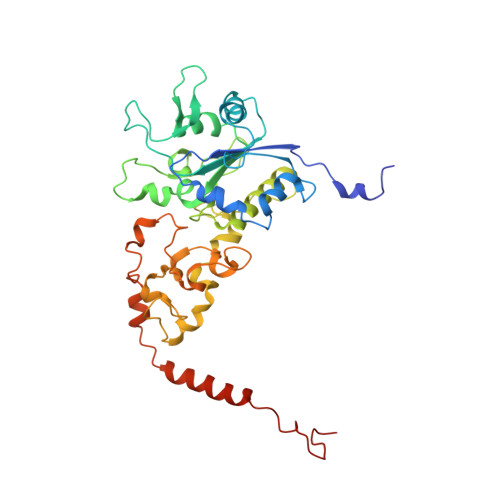
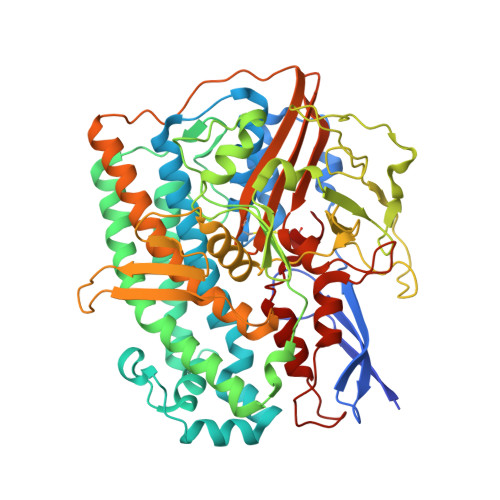
Structure of an Actinobacterial-Type [Nife]-Hydrogenase Reveals Insight Into O2-Tolerant H2 Oxidation.
Schafer, C., Bommer, M., Hennig, S.E., Jeoung, J., Dobbek, H., Lenz, O.(2016) Structure 24: 285
- PubMed: 26749450
- DOI: https://doi.org/10.1016/j.str.2015.11.010
- Primary Citation of Related Structures:
5AA5 - PubMed Abstract:
A novel group of bacterial [NiFe]-hydrogenases is responsible for high-affinity H2 uptake from the troposphere, and is therefore thought to play an important role in the global H2 cycle. Here we present the first crystal structure at 2.85-Å resolution of such an actinobacterial-type hydrogenase (AH), which was isolated from the dihydrogen oxidizing bacterium, Ralstonia eutropha. The enzyme has a dimeric structure carrying two active [NiFe] sites that are interconnected by six [4Fe4S] clusters over a range of approximately 90 Å. Unlike most other [NiFe]-hydrogenases, the [4Fe4S] cluster proximal to the [NiFe] site is coordinated by three cysteines and one aspartate. Mutagenesis experiments revealed that this aspartate residue is related to the apparent O2 insensitivity of the AH. Our data provide first structural insight into specialized hydrogenases that are supposed to consume atmospheric H2 under challenging conditions, i.e. at high O2 concentration and wide temperature and pH ranges.
- Institut für Biologie/Mikrobiologie, Humboldt-Universität zu Berlin, Chausseestraße 117, 10115 Berlin, Germany.
Organizational Affiliation: