Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein.
Gilman, M.S., Moin, S.M., Mas, V., Chen, M., Patel, N.K., Kramer, K., Zhu, Q., Kabeche, S.C., Kumar, A., Palomo, C., Beaumont, T., Baxa, U., Ulbrandt, N.D., Melero, J.A., Graham, B.S., McLellan, J.S.(2015) PLoS Pathog 11: e1005035-e1005035
- PubMed: 26161532
- DOI: https://doi.org/10.1371/journal.ppat.1005035
- Primary Citation of Related Structures:
4ZYK, 4ZYP - PubMed Abstract:
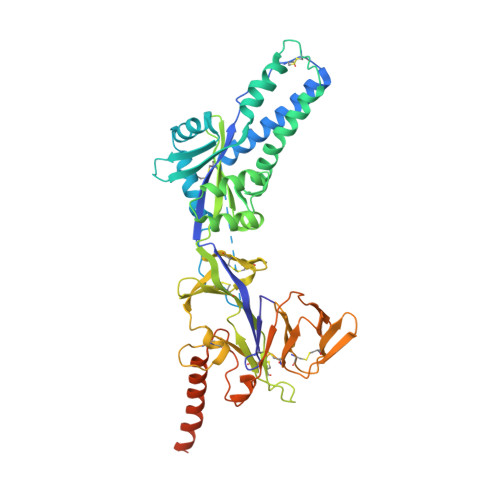
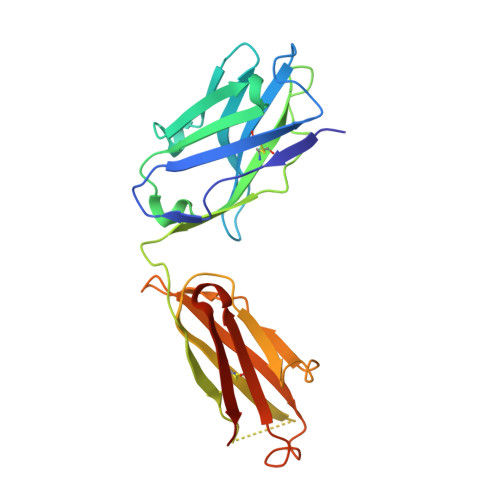
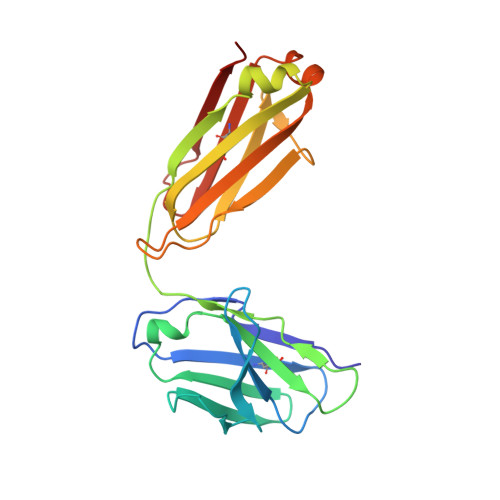
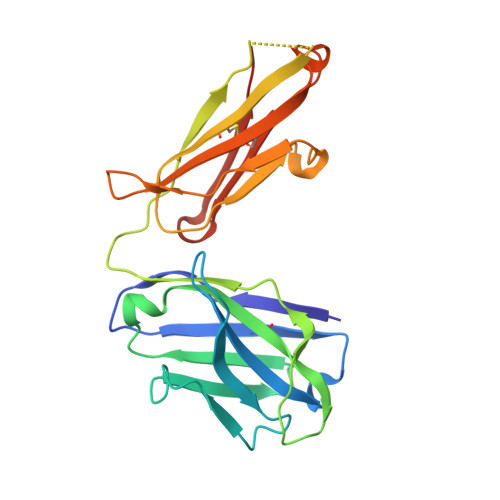
Prevention efforts for respiratory syncytial virus (RSV) have been advanced due to the recent isolation and characterization of antibodies that specifically recognize the prefusion conformation of the RSV fusion (F) glycoprotein. These potently neutralizing antibodies are in clinical development for passive prophylaxis and have also aided the design of vaccine antigens that display prefusion-specific epitopes. To date, prefusion-specific antibodies have been shown to target two antigenic sites on RSV F, but both of these sites are also present on monomeric forms of F. Here we present a structural and functional characterization of human antibody AM14, which potently neutralized laboratory strains and clinical isolates of RSV from both A and B subtypes. The crystal structure and location of escape mutations revealed that AM14 recognizes a quaternary epitope that spans two protomers and includes a region that undergoes extensive conformational changes in the pre- to postfusion F transition. Binding assays demonstrated that AM14 is unique in its specific recognition of trimeric furin-cleaved prefusion F, which is the mature form of F on infectious virions. These results demonstrate that the prefusion F trimer contains potent neutralizing epitopes not present on monomers and that AM14 should be particularly useful for characterizing the conformational state of RSV F-based vaccine antigens.
- Department of Biochemistry, Geisel School of Medicine at Dartmouth, Hanover, New Hampshire, United States of America.
Organizational Affiliation: