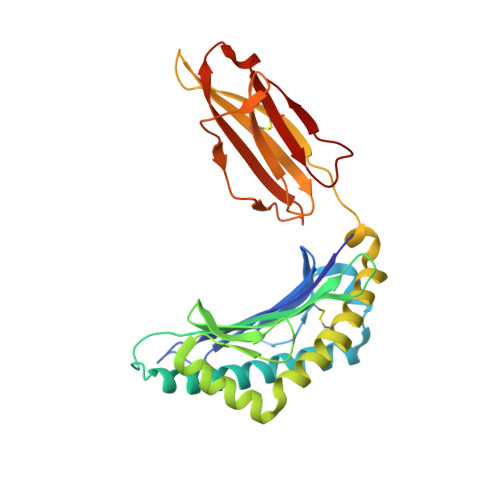
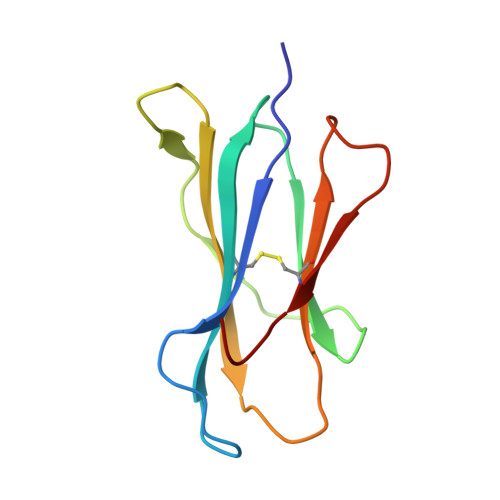
Structural Illumination of Equine MHC Class I Molecules Highlights Unconventional Epitope Presentation Manner That Is Evolved in Equine Leukocyte Antigen Alleles
Yao, S., Liu, J., Qi, J., Chen, R., Zhang, N., Liu, Y., Wang, J., Wu, Y., Gao, G.F., Xia, C.(2016) J Immunol 196: 1943-1954
- PubMed: 26764037
- DOI: https://doi.org/10.4049/jimmunol.1501352
- Primary Citation of Related Structures:
4ZUS, 4ZUT, 4ZUU, 4ZUV, 4ZUW - PubMed Abstract:
MHC class I (MHC I)-restricted virus-specific CTLs are implicated as critical components in the control of this naturally occurring lentivirus and in the protective immune response to the successfully applied attenuated equine infectious anemia virus vaccine in the horse. Nevertheless, the structural basis for how the equine MHC I presents epitope peptides remains unknown. In this study, we investigated the binding of several equine infectious anemia virus-derived epitope peptides by the ability to refold recombinant molecules and by thermal stability, and then by determining the x-ray structure of five peptide-MHC I complexes: equine MHC class I allele (Eqca)-N*00602/Env-RW12, Eqca-N*00602/Gag-GW12, Eqca-N*00602/Rev-QW11, Eqca-N*00602/Gag-CF9, and Eqca-N*00601/Gag-GW12. Although Eqca-N*00601 and Eqca-N*00602 differ by a single amino acid, Eqca-N*00601 exhibited a drastically different peptide presentation when binding a similar CTL epitope, Gag-GW12; the result makes the previously reported function clear to be non-cross-recognition between these two alleles. The structures plus Eqca-N*00602 complexed with a 9-mer peptide are particularly noteworthy in that we illuminated differences in apparent flexibility in the center of the epitope peptides for the complexes with Gag-GW12 as compared with Env-RW12, and a strict selection of epitope peptides with normal length. The featured preferences and unconventional presentations of long peptides by equine MHC I molecules provide structural bases to explain the exceptional anti-lentivirus immunity in the horse. We think that the beneficial reference points could serve as an initial platform for other human or animal lentiviruses.
- Department of Microbiology and Immunology, College of Veterinary Medicine, China Agricultural University, Beijing 100193, China; Department of Medical Microbiology and Immunology, University of Alberta, Edmonton, Alberta T6G 2R3, Canada;
Organizational Affiliation: