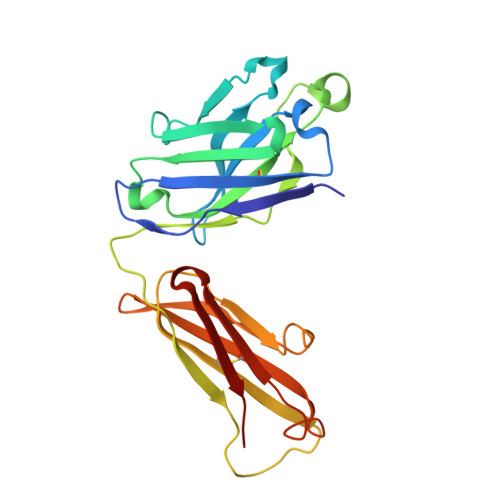
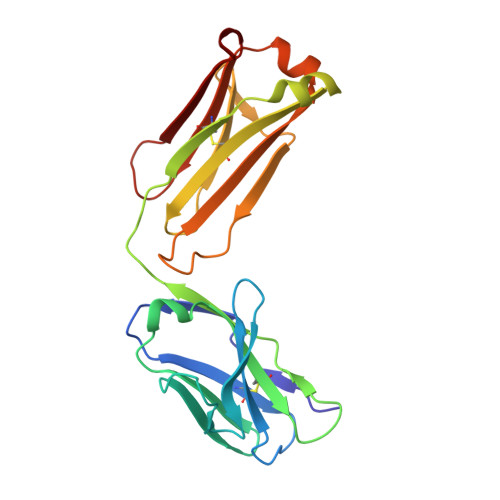
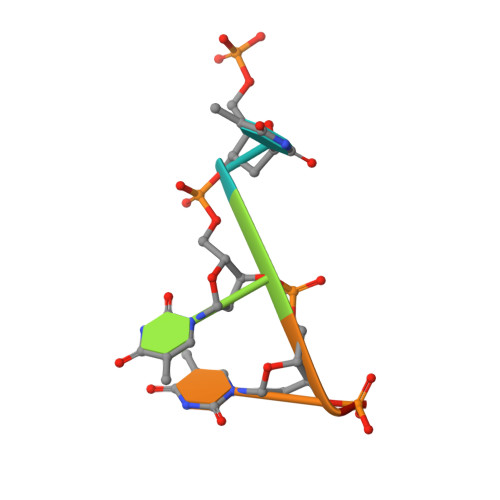
Structural Basis for Antibody Recognition of Lipid A: INSIGHTS TO POLYSPECIFICITY TOWARD SINGLE-STRANDED DNA.
Haji-Ghassemi, O., Muller-Loennies, S., Rodriguez, T., Brade, L., Kosma, P., Brade, H., Evans, S.V.(2015) J Biological Chem 290: 19629-19640
- PubMed: 26085093
- DOI: https://doi.org/10.1074/jbc.M115.657874
- Primary Citation of Related Structures:
4ODS, 4ODT, 4ODU, 4ODV, 4ODW, 4Z8F, 4Z95 - PubMed Abstract:
Septic shock is a leading cause of death, and it results from an inflammatory cascade triggered by the presence of microbial products in the blood. Certain LPS from Gram-negative bacteria are very potent inducers and are responsible for a high percentage of septic shock cases. Despite decades of research, mAbs specific for lipid A (the endotoxic principle of LPS) have not been successfully developed into a clinical treatment for sepsis. To understand the molecular basis for the observed inability to translate in vitro specificity for lipid A into clinical potential, the structures of antigen-binding fragments of mAbs S1-15 and A6 have been determined both in complex with lipid A carbohydrate backbone and in the unliganded form. The two antibodies have separate germ line origins that generate two markedly different combining-site pockets that are complementary both in shape and charge to the antigen. mAb A6 binds lipid A through both variable light and heavy chain residues, whereas S1-15 utilizes exclusively the variable heavy chain. Both antibodies bind lipid A such that the GlcN-O6 attachment point for the core oligosaccharide is buried in the combining site, which explains the lack of LPS recognition. Longstanding reports of polyspecificity of anti-lipid A antibodies toward single-stranded DNA combined with observed homology of S1-15 and A6 and the reports of several single-stranded DNA-specific mAbs prompted the determination of the structure of S1-15 in complex with single-stranded DNA fragments, which may provide clues about the genesis of autoimmune diseases such as systemic lupus erythematosus, thyroiditis, and rheumatic autoimmune diseases.
- From the Department of Biochemistry and Microbiology, University of Victoria, Victoria, British Columbia V8P 3P6, Canada.
Organizational Affiliation: