Structural basis of cucumisin protease activity regulation by its propeptide
Sotokawauchi, A., Kato-Murayama, M., Murayama, K., Hosaka, T., Maeda, I., Onjo, M., Ohsawa, N., Kato, D.I., Arima, K., Shirouzu, M.(2017) J Biochem 161: 45-53
- PubMed: 27616715
- DOI: https://doi.org/10.1093/jb/mvw053
- Primary Citation of Related Structures:
4YN3 - PubMed Abstract:
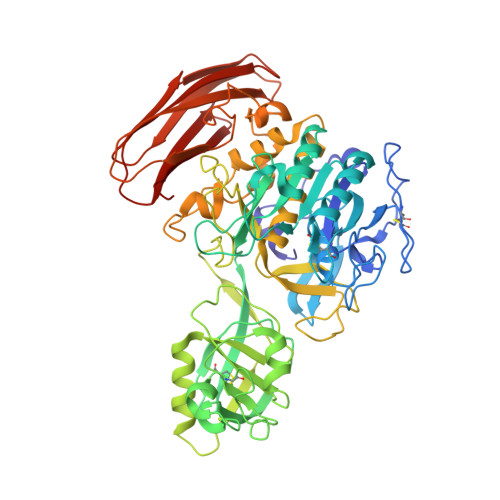
Cucumisin [EC 3.4.21.25], a subtilisin-like serine endopeptidase, was isolated from melon fruit, Cucumis melo L. Mature cucumisin (67 kDa, 621 residues) is produced by removal of the propeptide (10 kDa, 88 residues) from the cucumisin precursor by subsequence processing. It is reported that cucumisin is inhibited by its own propeptide. The crystal structure of mature cucumisin is reported to be composed of three domains: the subtilisin-like catalytic domain, the protease-associated domain and the C-terminal fibronectin-III-like domain. In this study, the crystal structure of the mature cucumisin•propeptide complex was determined by the molecular replacement method and refined at 1.95 Å resolution. In this complex, the propeptide had a domain of the α-β sandwich motif with four-stranded antiparallel β-sheets, two helices and a strand of the C-terminal region. The β-sheets of the propeptide bind to two parallel surface helices of cucumisin through hydrophobic interaction and 27 hydrogen bonds. The C-terminus of the propeptide binds to the cleft of the active site as peptide substrates. The inhibitory assay suggested that the C-terminal seven residues of the propeptide do not inhibit the cucumisin activity. The crystal structure of the cucumisin•propeptide complex revealed the regulation mechanism of cucumisin activity.
- Department of Chemistry and Bioscience, Faculty of Science, Kagoshima University Graduate School of Science and Engineering, Kagoshima 890-0065, Japan.
Organizational Affiliation: