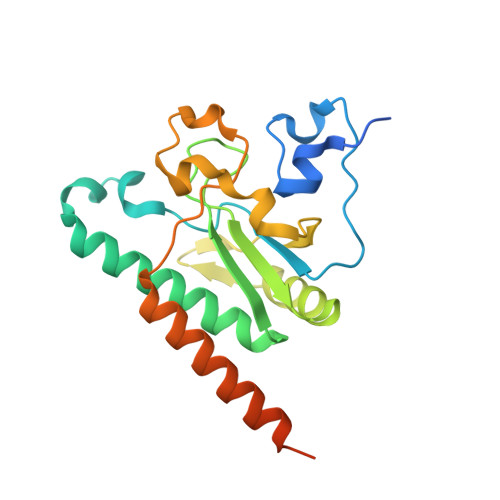
A novel 3' splice site recognition by the two zinc fingers in the U2AF small subunit.
Yoshida, H., Park, S.Y., Oda, T., Akiyoshi, T., Sato, M., Shirouzu, M., Tsuda, K., Kuwasako, K., Unzai, S., Muto, Y., Urano, T., Obayashi, E.(2015) Genes Dev 29: 1649-1660
- PubMed: 26215567
- DOI: https://doi.org/10.1101/gad.267104.115
- Primary Citation of Related Structures:
4YH8 - PubMed Abstract:
The pre-mRNA splicing reaction of eukaryotic cells has to be carried out extremely accurately, as failure to recognize the splice sites correctly causes serious disease. The small subunit of the U2AF heterodimer is essential for the determination of 3' splice sites in pre-mRNA splicing, and several single-residue mutations of the U2AF small subunit cause severe disorders such as myelodysplastic syndromes. However, the mechanism of RNA recognition is poorly understood. Here we solved the crystal structure of the U2AF small subunit (U2AF23) from fission yeast, consisting of an RNA recognition motif (RRM) domain flanked by two conserved CCCH-type zinc fingers (ZFs). The two ZFs are positioned side by side on the β sheet of the RRM domain. Further mutational analysis revealed that the ZFs bind cooperatively to the target RNA sequence, but the RRM domain acts simply as a scaffold to organize the ZFs and does not itself contact the RNA directly. This completely novel and unexpected mode of RNA-binding mechanism by the U2AF small subunit sheds light on splicing errors caused by mutations of this highly conserved protein.
- Graduate School of Medical Life Science, Yokohama City University, Tsurumi-ku, Yokohama 230-0045, Japan; Drug Design Group, Kanagawa Academy of Science and Technology (KAST), Takatsu-ku, Kawasaki 213-0012, Japan;
Organizational Affiliation: