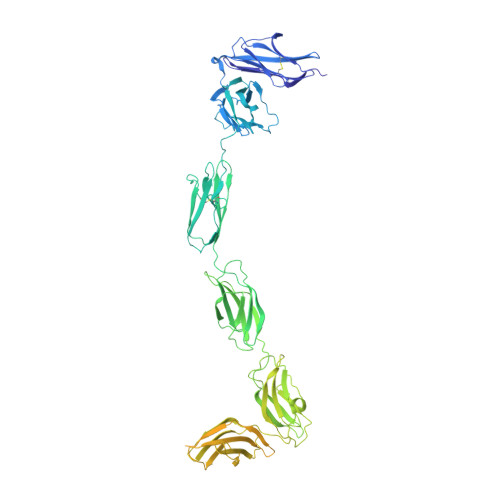
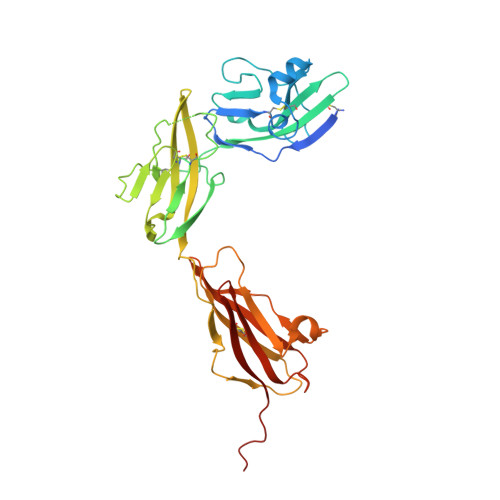
Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Yamagata, A., Yoshida, T., Sato, Y., Goto-Ito, S., Uemura, T., Maeda, A., Shiroshima, T., Iwasawa-Okamoto, S., Mori, H., Mishina, M., Fukai, S.(2015) Nat Commun 6: 6926-6926
- PubMed: 25908590
- DOI: https://doi.org/10.1038/ncomms7926
- Primary Citation of Related Structures:
4YFD, 4YFE, 4YFG, 4YH6, 4YH7, 5Y32 - PubMed Abstract:
Synapse formation is triggered through trans-synaptic interaction between pairs of pre- and postsynaptic adhesion molecules, the specificity of which depends on splice inserts known as 'splice-insert signaling codes'. Receptor protein tyrosine phosphatase δ (PTPδ) can bidirectionally induce pre- and postsynaptic differentiation of neurons by trans-synaptically binding to interleukin-1 receptor accessory protein (IL-1RAcP) and IL-1RAcP-like-1 (IL1RAPL1) in a splicing-dependent manner. Here, we report crystal structures of PTPδ in complex with IL1RAPL1 and IL-1RAcP. The first immunoglobulin-like (Ig) domain of IL1RAPL1 directly recognizes the first splice insert, which is critical for binding to IL1RAPL1. The second splice insert functions as an adjustable linker that positions the Ig2 and Ig3 domains of PTPδ for simultaneously interacting with the Ig1 domain of IL1RAPL1 or IL-1RAcP. We further identified the IL1RAPL1-specific interaction, which appears coupled to the first-splice-insert-mediated interaction. Our results thus reveal the decoding mechanism of splice-insert signaling codes for synaptic differentiation induced by trans-synaptic adhesion between PTPδ and IL1RAPL1/IL-1RAcP.
- 1] Structural Biology Laboratory, Life Science Division, Synchrotron Radiation Research Organization and Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo 113-0032, Japan [2] Department of Medical Genome Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Chiba 277-8501, Japan [3] CREST, JST, Saitama 332-0012, Japan.
Organizational Affiliation: