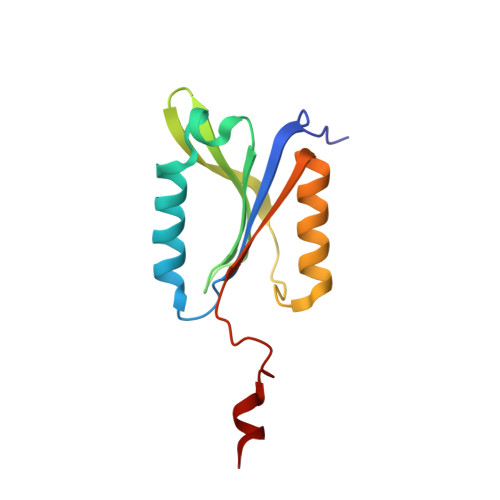
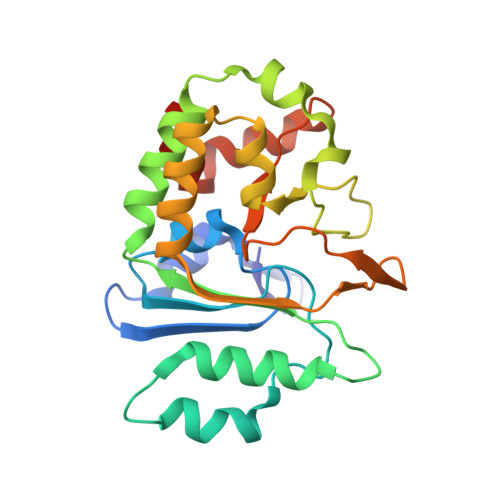
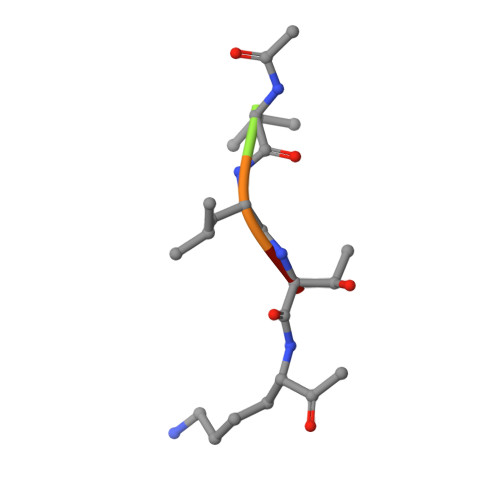
Substrate Profiling and High Resolution Co-complex Crystal Structure of a Secreted C11 Protease Conserved across Commensal Bacteria.
Roncase, E.J., Moon, C., Chatterjee, S., Gonzalez-Paez, G.E., Craik, C.S., O'Donoghue, A.J., Wolan, D.W.(2017) ACS Chem Biol 12: 1556-1565
- PubMed: 28414448
- DOI: https://doi.org/10.1021/acschembio.7b00143
- Primary Citation of Related Structures:
4YEC - PubMed Abstract:
Cysteine proteases are among the most abundant hydrolytic enzymes produced by bacteria, and this diverse family of proteins have significant biological roles in bacterial viability and environmental interactions. Members of the clostripain-like (C11) family of cysteine proteases from commensal gut bacterial strains have recently been shown to mediate immune responses by inducing neutrophil phagocytosis and activating bacterial pathogenic toxins. Development of substrates, inhibitors, and probes that target C11 proteases from enteric bacteria will help to establish the role of these proteins at the interface of the host and microbiome in health and disease. We employed a mass spectrometry-based substrate profiling method to identify an optimal peptide substrate of PmC11, a C11 protease secreted by the commensal bacterium Parabacteroides merdae. Using this substrate sequence information, we synthesized a panel of fluorogenic substrates to calculate k cat and K M and to evaluate the importance of the P2 amino acid for substrate turnover. A potent and irreversible tetrapeptide inhibitor with a C-terminal acyloxymethyl ketone warhead, Ac-VLTK-AOMK, was then synthesized. We determined the crystal structure of PmC11 in complex with this inhibitor and uncovered key active-site interactions that govern PmC11 substrate recognition and specificity. This is the first C11 protease structure in complex with a substrate mimetic and is also the highest resolution crystal structure of a C11 protease to date at 1.12 Å resolution. Importantly, subjecting human epithelial cell lysates to PmC11 hydrolysis in combination with subtiligase-based N-terminal labeling and tandem mass spectrometry proteomics complemented the stringent substrate specificity observed in the in vitro substrate profiling experiment. The combination of chemical biological, biophysical, and biochemical techniques presented here to elucidate and characterize PmC11 substrate selectivity can be expanded to other proteases and the development of chemical tools to study these essential proteins in biologically relevant samples, such as the highly complex distal gut microbiome.
- Department of Molecular Medicine, The Scripps Research Institute , 10550 North Torrey Pines Road, La Jolla, California 92037, United States.
Organizational Affiliation: