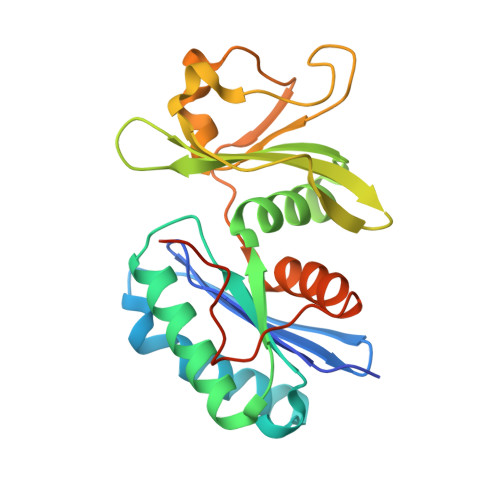
Crystal structure of YeaZ from Pseudomonas aeruginosa.
Vecchietti, D., Ferrara, S., Rusmini, R., Macchi, R., Milani, M., Bertoni, G.(2016) Biochem Biophys Res Commun 470: 460-465
- PubMed: 26768361
- DOI: https://doi.org/10.1016/j.bbrc.2016.01.008
- Primary Citation of Related Structures:
4Y0W - PubMed Abstract:
The Pseudomonas aeruginosa PA3685 locus encodes a conserved protein that shares 49% sequence identity with Escherichia coli YeaZ, which was recently reported as involved in the biosynthesis of threonylcarbamoyl adenosine (t(6)A), a universal modified tRNA nucleoside. Many YeaZ orthologues were reported as "essential for life" among various bacterial species, suggesting a critical role for both these proteins and for the t(6)A biosynthetic pathway. We provide here evidences that PA3685 protein (PaYeaZ) is essential. Additionally, we describe its purification, crystallization, and crystallographic structure. The crystal structure shows that PaYeaZ is composed of two domains one of which is the platform to form protein-protein interaction involved either in homodimeric assembly or in the formation of the multiprotein complex required for the synthesis of t(6)A. These features make the PaYeaZ protein a potential target candidate for the design of novel inhibitors able to hinder the complex formation and expected to abolish the crucial activity of t(6)A synthesis.
- Dipartimento di Bioscienze, Università di Milano, Via Celoria 26, I-20133, Milano, Italy.
Organizational Affiliation: