Structural insights into the binding of the human receptor for advanced glycation end products (RAGE) by S100B, as revealed by an S100B-RAGE-derived peptide complex.
Jensen, J.L., Indurthi, V.S., Neau, D.B., Vetter, S.W., Colbert, C.L.(2015) Acta Crystallogr D Biol Crystallogr 71: 1176-1183
- PubMed: 25945582
- DOI: https://doi.org/10.1107/S1399004715004216
- Primary Citation of Related Structures:
4XYN - PubMed Abstract:
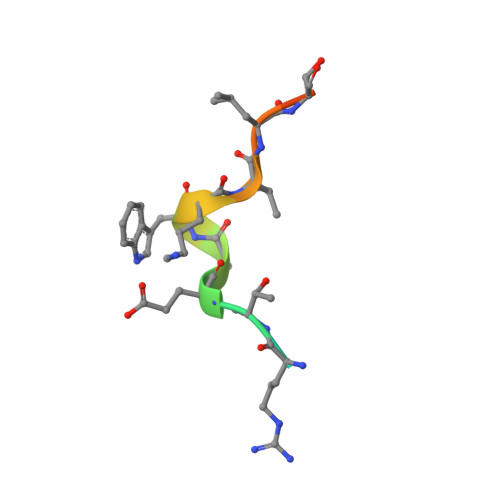
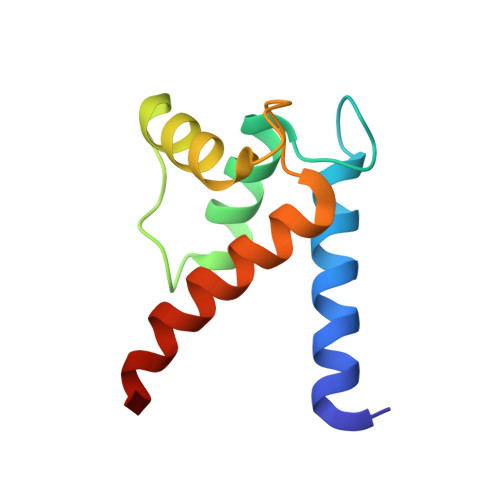
S100B is a damage-associated molecular pattern protein that, when released into the extracellular milieu, triggers initiation of the inflammatory response through the receptor for advanced glycation end products (RAGE). Recognition of S100B is accomplished via the amino-terminal variable immunoglobulin domain (V-domain) of RAGE. To gain insights into this interaction, a complex between S100B and a 15-amino-acid peptide derived from residues 54-68 of the V-domain was crystallized. The X-ray crystal structure was solved to 2.55 Å resolution. There are two dimers of S100B and one peptide in the asymmetric unit. The binding interface of this peptide is compared with that found in the complex between S100B and the 12-amino-acid CapZ-derived peptide TRTK-12. This comparison reveals that although the peptides adopt completely different backbone structures, the residues buried at the interface interact with S100B in similar regions to form stable complexes. The binding affinities of S100B for the intact wild-type V-domain and a W61A V-domain mutant were determined to be 2.7 ± 0.5 and 1.3 ± 0.7 µM, respectively, using fluorescence titration experiments. These observations lead to a model whereby conformational flexibility in the RAGE receptor allows the adoption of a binding conformation for interaction with the stable hydrophobic groove on the surface of S100B.
- Department of Chemistry and Biochemistry, North Dakota State University, PO Box 6050, Fargo, ND 58108-6050, USA.
Organizational Affiliation: