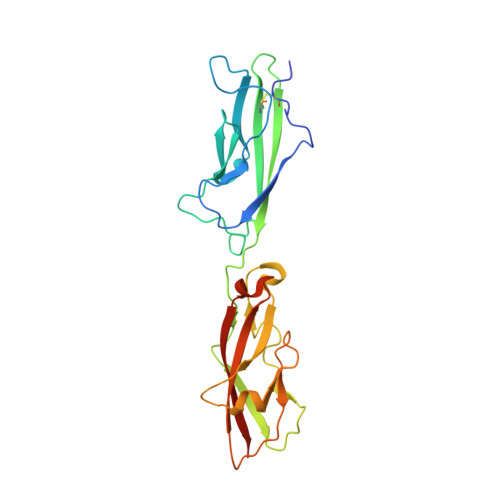
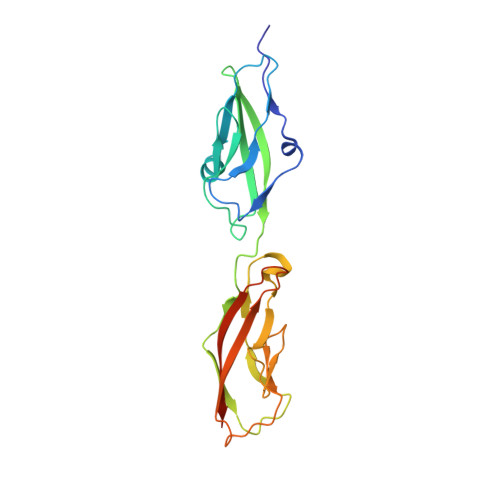
Tuning Inner-Ear Tip-Link Affinity Through Alternatively Spliced Variants of Protocadherin-15.
Narui, Y., Sotomayor, M.(2018) Biochemistry 57: 1702-1710
- PubMed: 29443515
- DOI: https://doi.org/10.1021/acs.biochem.7b01075
- Primary Citation of Related Structures:
4XXW - PubMed Abstract:
Human hearing relies upon the tip-to-tip interaction of two nonclassical cadherins, protocadherin-15 (PCDH15) and cadherin-23 (CDH23). Together, these proteins form a filament called the tip link that connects neighboring stereocilia of mechanosensitive hair cells. As sound waves enter the cochlea, the stereocilia deflect and tension is applied to the tip link, opening nearby transduction channels. Disruption of the tip link by loud sound or calcium chelators eliminates transduction currents and illustrates that tip-link integrity is critical for mechanosensing. Tip-link remodeling after disruption is a dynamic process, which can lead to the formation of atypical complexes that incorporate alternatively spliced variants of PCDH15. These variants are categorized into six groups (N1-N6) based upon differences in the first two extracellular cadherin (EC) repeats. Here, we characterized the two N-terminal EC repeats of all PCDH15 variants (pcdh15(N1) to pcdh15(N6)) and combined these variants to test complex formation. We solved the crystal structure of a new complex composed of CDH23 EC1-2 (cdh23) and pcdh15(N2) at 2.3 Å resolution and compared it to the canonical cdh23-pcdh15(N1) complex. While there were subtle structural differences, the binding affinity between cdh23 and pcdh15(N2) is ∼6 times weaker than cdh23 and pcdh15(N1) as determined by surface plasmon resonance analysis. Steered molecular dynamics simulations predict that the unbinding force of the cdh23-pcdh15(N2) complex can be lower than the canonical tip link. Our results demonstrate that alternative heterophilic tip-link structures form stable protein-protein interactions in vitro and suggest that homophilic PCDH15-PCDH15 tip links form through the interaction of additional EC repeats.
- Department of Chemistry and Biochemistry , The Ohio State University , Columbus , Ohio 43210 , United States.
Organizational Affiliation: