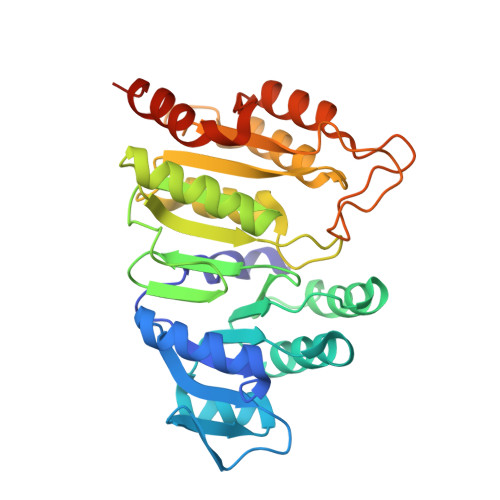
Structure of GTP-specific succinyl-CoA synthetase in complex with CoA.
Huang, J., Malhi, M., Deneke, J., Fraser, M.E.(2015) Acta Crystallogr F Struct Biol Commun 71: 1067-1071
- PubMed: 26249701
- DOI: https://doi.org/10.1107/S2053230X15011188
- Primary Citation of Related Structures:
4XX0 - PubMed Abstract:
Pig GTP-specific succinyl-CoA synthetase is an αβ-heterodimer. The crystal structure of the complex with the substrate CoA was determined at 2.1 Å resolution. The structure shows CoA bound to the amino-terminal domain of the α-subunit, with the free thiol extending from the adenine portion into the site where the catalytic histidine residue resides.
- Department of Biological Sciences, University of Calgary, 2500 University Drive NW, Calgary, AB T2N 1N4, Canada.
Organizational Affiliation: