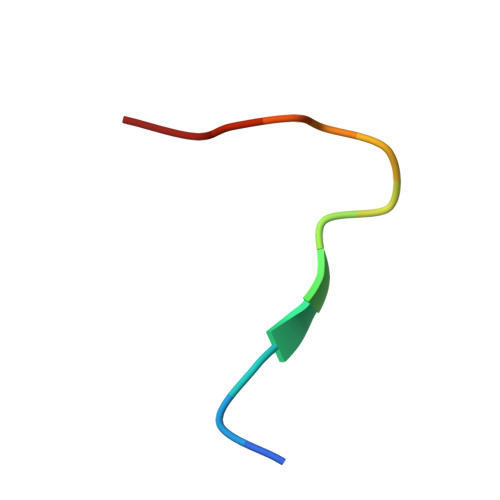
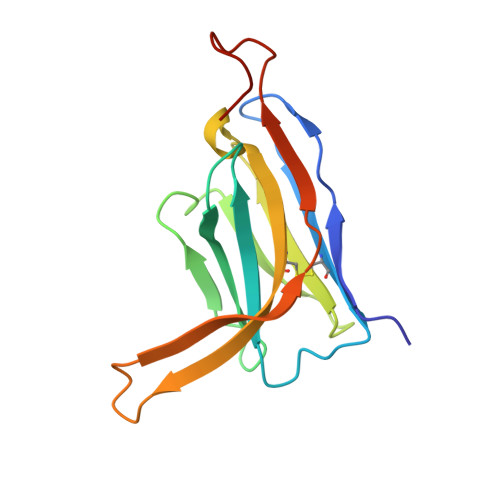
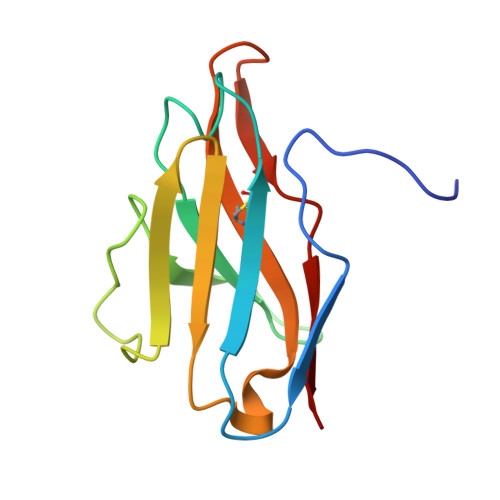
Structural basis for penetration of the glycan shield of hepatitis C virus e2 glycoprotein by a broadly neutralizing human antibody.
Li, Y., Pierce, B.G., Wang, Q., Keck, Z.Y., Fuerst, T.R., Foung, S.K., Mariuzza, R.A.(2015) J Biological Chem 290: 10117-10125
- PubMed: 25737449
- DOI: https://doi.org/10.1074/jbc.M115.643528
- Primary Citation of Related Structures:
4XVJ - PubMed Abstract:
Hepatitis C virus (HCV) is a major cause of liver cirrhosis and hepatocellular carcinoma. A challenge for HCV vaccine development is to identify conserved epitopes able to elicit protective antibodies against this highly diverse virus. Glycan shielding is a mechanism by which HCV masks such epitopes on its E2 envelope glycoprotein. Antibodies to the E2 region comprising residues 412-423 (E2(412-423)) have broadly neutralizing activities. However, an adaptive mutation in this linear epitope, N417S, is associated with a glycosylation shift from Asn-417 to Asn-415 that enables HCV to escape neutralization by mAbs such as HCV1 and AP33. By contrast, the human mAb HC33.1 can neutralize virus bearing the N417S mutation. To understand how HC33.1 penetrates the glycan shield created by the glycosylation shift to Asn-415, we determined the structure of this broadly neutralizing mAb in complex with its E2(412-423) epitope to 2.0 Å resolution. The conformation of E2(412-423) bound to HC33.1 is distinct from the β-hairpin conformation of this peptide bound to HCV1 or AP33, because of disruption of the β-hairpin through interactions with the unusually long complementarity-determining region 3 of the HC33.1 heavy chain. Whereas Asn-415 is buried by HCV1 and AP33, it is solvent-exposed in the HC33.1-E2(412-423) complex, such that glycosylation of Asn-415 would not prevent antibody binding. Furthermore, our results highlight the structural flexibility of the E2(412-423) epitope, which may serve as an immune evasion strategy to impede induction of antibodies targeting this site by reducing its antigenicity.
- From the University of Maryland Institute for Bioscience and Biotechnology Research, W. M. Keck Laboratory for Structural Biology, Rockville, Maryland 20850, the Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, Maryland 20742, and.
Organizational Affiliation: